We provide functionality for writing a Tree to different file formats that allow visualization via external tree viewers:
newick tree files that can be read and visualized by most other tools.phyloxml tree files, best suited for the Archaeopteryx tree viewer.nexus files, which internally also use newick to store trees; best suited for the FigTree tree viewer.svg files, yielding publication quality vector graphics. This is the only type of visualization that directly supports to also draw a legend for edge colors.Remarks: See Czech et al. 2017 for a comprehensive review on tree viewers.
In the following, we assume that the following namespaces are available:
Many of the tree visualization that we for example explain in the Placement Analyses tutorial or that are part of our analysis program gappa visualize trees with individually colored edges. Here, we explain how this is done in genesis.
Firstly, get a tree and see how many edges it has:
Now, create a vector of Color objects for each edge of the tree:
See the Colors tutorial on how color is handled in genesis.
In the above snippet, we made all edges red. The TreeEdge::index() of each edge corresponds to one position in the vector. See the Indices of Links, Nodes, and Edges section for details on indices. One can change edge colors in the vector according to these indices. For example, make all edges leading to tip nodes blue:
In an actual algorithm for analyzing some data, the colors can then be set as needed, for example by using a color gradient. Color gradients can be achieved using ColorMap and ColorNormalization, see the Colors tutorial for details.
Now that we have a std::vector of Colors for each edge of the tree, we can store the result in various formats.
As discussed in the sections Reading and Writing and Reading and Writing Revisited, one can use NewickWriter and PhyloxmlWriter to write Trees to newick and phyloxml files.
The newick format does not support colors. However, for visualizing just the tree topology with taxon names and branch lengths, it is still useful. If just these defaults are needed, we offer a simple function that wraps the CommonTreeNewickWriter:
This can be visualized with most tree viewers.
The phyloxml format offers some more annotations, and allows for color annotations for its branches. This is for example used by the guppy tool that is part of the pplacer suite of programs of phylogenetic placement. See their notes on phyloxml for some details. See phyloxml.org for details on the format.
To write a tree with colored edges, we can again use a simple wrapper function:
The resulting phyloxml file is best viewed with the Archaeopteryx tree viewer. There is also write_tree_to_phyloxml_file() as a wrapper without colors, as well as a version that takes a list of per-edge values and a ColorMap and ColorNormalization, and performs the transformation of values into colors as well. See their implementation to get the gist of how this works behind the scenes.
The nexus format is a container format that also allows to store trees. See for example this page for some details on the format.
It works similar to the phyloxml wrappers:
The resulting file is best viewed with the FigTree tree viewer. Again, there is write_tree_to_nexus_file() for a version without colors, and one that also transforms per-edge values to colors first. See the implementation of these wrapper functions to understand how to build a NexusDocument containing a tree from scratch.
The above functions yield files that can be viewed with external tree viewers. This is cumbersome for automatization and for testing purposes. Furthermore, tree viewers do not support advanced features such as putting a legend for the color scale next to the tree. Hence, we offer to draw trees to svg files directly.
This means that all layout parameters have to be set in the code. Most importantly, these are:
These properties are summarized in LayoutParameters, which can be used again with simple wrapper functions to get svg drawings of a tree. We here use the edge_colors from above to show edges that lead to leaf nodes in blue:
This yields the following figure:

Again, there also is a version write_tree_to_svg_file() that does not use colors, but simply write a tree with black branches.
Writing to svg is the only way to directly draw the legend for the edge colors into the document. This allows to interpret which edge color corresponds to which value. Let's visualize the branch lengths of the tree:
See the Colors tutorial on how color is handled in genesis, and on the ColorMap and ColorNormalization classes in particular.
The above snippet yields a figure similar to the following one, where longer branches have a darker color:
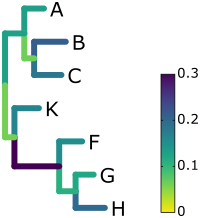
The figure was fine tuned afterwards, because programmatically setting all font sizes, line widths, spaces between figure elements, etc, to nice values is tricky. We however did not remove or add any elements to the drawing: For example, all parts of the legend, including the labeling with fitting intervals (0 to 0.3), are automatic. We recommend Inkscape for fine tuning of the resulting svg files.
The above functions are all wrappers for simplicity. They cover the most common use cases, and in particular cover per-edge colors. They however do not allow for other types of individual edge styles (dashed, line widths, etc). If this is required, one can use the implementation of the wrappers as a starting point to create their own visualizations.
The LayoutBase class is a good starting point for more specialized visualizations. It allows:
A final set of functions that we want to mention are the heat_tree() functions, which allow to draw a tree to svg, with a heat matrix next to it for each of its taxa, which can for example show per-sample data for each of the taxa of the tree. This is commonly used in metagenomic and environmental studies.