Classes | |
| class | AttributeTreeEdgeData |
| Data class for AttributeTreeEdges. More... | |
| class | AttributeTreeNodeData |
| Data class for AttributeTreeNodes. More... | |
| struct | BalanceData |
| Data needed to calculate balances. More... | |
| struct | BalanceSettings |
| Settings to calculate balances and the Phylogenetic ILR Transform. More... | |
| class | BaseEdgeData |
| Base class for storing data on Edges of a Tree. More... | |
| class | BaseNodeData |
| Base class for storing data on Nodes of a Tree. More... | |
| class | Bipartition |
| class | CircularLayout |
| class | ColorWriterPlugin |
| Base class for creating plugin classes that allow coloring of Tree edges. More... | |
| class | CommonEdgeData |
| Common class containing the commonly needed data for tree edges. More... | |
| class | CommonNodeData |
| Common class containing the commonly needed data for tree nodes. More... | |
| class | CommonTreeNewickReader |
| Read default Newick trees, i.e., trees with names and branch lengths. More... | |
| class | CommonTreeNewickReaderPlugin |
| Provide a set of plugin functions for NewickReader to read a CommonTree. More... | |
| class | CommonTreeNewickWriter |
| Write default Newick trees, i.e., trees with names and branch lengths. More... | |
| class | CommonTreeNewickWriterPlugin |
| Provide a set of plugin functions for NewickWriter to write a CommonTree. More... | |
| class | CommonTreePhyloxmlWriter |
| class | CommonTreePhyloxmlWriterPlugin |
| struct | HeatTreeParameters |
| class | IndexedAttributeTreeNewickReader |
| Read Newick trees with ordered attributes for the Nodes and Edges. More... | |
| class | IndexedAttributeTreeNewickReaderPlugin |
| Provide a set of plugin functions for NewickReader to read ordered attributes of the Nodes and Edges into an AttributeTree. More... | |
| class | IteratorEulertour |
| class | IteratorLevelorder |
| class | IteratorNodeLinks |
| class | IteratorPath |
| class | IteratorPathSet |
| Iterate the path between two TreeNodes (non-linearily), given their lowest common ancestor (LCA). More... | |
| class | IteratorPostorder |
| class | IteratorPreorder |
| class | KeyedAttributeTreeNewickReader |
| Read default Newick trees, i.e., trees with names and branch lengths. More... | |
| class | KeyedAttributeTreeNewickReaderPlugin |
| Provide a set of plugin functions for NewickReader to read key-value-pair data attributes into an AttributeTree. More... | |
| class | LayoutBase |
| class | LayoutEdgeData |
| Data class for LayoutTreeEdges. More... | |
| class | LayoutNodeData |
| Data class for LayoutTreeNodes. More... | |
| struct | LayoutParameters |
| Collection of params for simple tree to svg wrapper functions. More... | |
| class | LcaLookup |
| Fast lookup of the lowest common ancestor (LCA) of two TreeNodes, relative to an arbitrary root node. More... | |
| class | MassTreeEdgeData |
| Data class for MassTreeEdges. Stores the branch length and a list of masses with their positions along the edge. More... | |
| class | MassTreeKmeans |
| class | MassTreeNodeData |
| Data class for MassTreeNodes. Stores taxon names. More... | |
| class | NewickBroker |
| Stores a Newick tree in an intermediate format that can be further processed into a Tree. More... | |
| struct | NewickBrokerElement |
| Store the information for one element of a Newick tree. More... | |
| class | NewickColorWriterPlugin |
| Plugin class for Newick output that allows coloring of edges. More... | |
| class | NewickInputIterator |
| Iterate an input stream and parse it as Newick trees. More... | |
| class | NewickReader |
| class | NewickWriter |
| Write a Tree to Newick format. More... | |
| struct | PhyloFactor |
| A single phylogenetic factor. More... | |
| struct | PhyloFactorCladeColors |
| Store a set of colors for making visualizations of the clades of all phylo factors. More... | |
| struct | PhyloFactorSingleColors |
| Store a set of colors for making visualizations of individual phylo factors. More... | |
| class | PhyloxmlColorWriterPlugin |
| Plugin class for PhyloXML output that allows coloring of edges. More... | |
| class | PhyloxmlWriter |
| Write a Tree to Phyloxml format. More... | |
| class | PrinterCompact |
| Print a Tree in a compact form, i.e., each node and edge on one line. More... | |
| class | PrinterDetailed |
| Print a text representation of the Tree, showing all nodes, edges and links with their indices. More... | |
| class | PrinterTable |
| Print lists of all nodes, edges and links including their indices and connections with each other. More... | |
| class | RectangularLayout |
| class | SimpleNewickEdgeData |
| Data class for SimpleNewickTreeEdges. More... | |
| class | SimpleNewickNodeData |
| Data class for SimpleNewickTreeNodes. More... | |
| class | SimpleNewickTreeNewickReader |
| class | SimpleNewickTreeNewickReaderPlugin |
| class | SimpleNewickTreeNewickWriter |
| class | SimpleNewickTreeNewickWriterPlugin |
| class | SquashClustering |
| Perform Squash Clustering. More... | |
| class | Subtree |
| Reference to a subtree of a Tree. More... | |
| class | Tree |
| Class for representing phylogenetic trees. More... | |
| class | TreeEdge |
| class | TreeLink |
| class | TreeNode |
| class | TreeSet |
Functions | |
| TreeNode & | add_new_leaf_node (Tree &tree, TreeEdge &target_edge, std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> adjust_edges={}) |
| Add a new Node as a leaf to an existing Edge, by also adding a new Node in the middle of that Edge. More... | |
| TreeNode & | add_new_node (Tree &tree, TreeEdge &target_edge, std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> adjust_edges={}) |
| Add a new Node that splits an existing Edge. More... | |
| TreeNode & | add_new_node (Tree &tree, TreeNode &target_node) |
| Add a new Node as a leaf to an existing Node. More... | |
| Tree | average_branch_length_tree (std::vector< Tree > const &tset) |
| Return a Tree where the branch lengths are the average of the Trees in the given vector of Trees or TreeSet, given that they all have the same topology. More... | |
| bool | belongs_to (Subtree const &subt, Tree const &tree) |
| Return whether the Subtree belongs to the Tree, i.e., whether its link is owned by the Tree. More... | |
| bool | belongs_to (Tree const &tree, Subtree const &subt) |
| Return whether the Subtree belongs to the Tree, i.e., whether its link is owned by the Tree. More... | |
| bool | belongs_to (Tree const &tree, TreeEdge const &edge) |
| Return whether the TreeEdge belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| bool | belongs_to (Tree const &tree, TreeLink const &link) |
| Return whether the TreeLink belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| bool | belongs_to (Tree const &tree, TreeNode const &node) |
| Return whether the TreeNode belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| bool | belongs_to (TreeEdge const &edge, Tree const &tree) |
| Return whether the TreeEdge belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| bool | belongs_to (TreeLink const &link, Tree const &tree) |
| Return whether the TreeLink belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| bool | belongs_to (TreeNode const &node, Tree const &tree) |
| Return whether the TreeNode belongs to the Tree, i.e., whether it is owned by the Tree. More... | |
| std::vector< Bipartition > | bipartition_set (Tree const &tree) |
| std::vector< double > | branch_lengths (Tree const &tree) |
| Get a vector of all branch lengths of a Tree, index by the edge index. More... | |
| void | change_rooting (Tree &tree, TreeLink &at_link) |
| "Reroot" the Tree at the given TreeLink. More... | |
| void | change_rooting (Tree &tree, TreeNode &at_node) |
| Reroot the Tree at the given TreeNode. More... | |
| std::vector< std::pair< TreeNode const *, size_t > > | closest_leaf_depth_vector (Tree const &tree) |
| Returns a vector containing the closest leaf node for each node, measured in number of edges between them and its depth (number of edges between them). More... | |
| std::vector< std::pair< TreeNode const *, double > > | closest_leaf_distance_vector (Tree const &tree) |
| Return a vector containing the closest leaf node for each node, using the branch_length as distance measure. More... | |
| std::vector< std::pair< TreeNode const *, double > > | closest_leaf_distance_vector (Tree const &tree, utils::Matrix< double > const &node_distances) |
| Tree | convert (Tree const &source, std::function< std::unique_ptr< BaseNodeData >(BaseNodeData const &node_data)> node_data_converter, std::function< std::unique_ptr< BaseEdgeData >(BaseEdgeData const &edge_data)> edge_data_converter) |
| Create a tree with the same topology as the source tree, while converting its data. More... | |
| AttributeTree | convert_common_tree_to_attribute_tree (CommonTree const &source) |
| Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an AttributeTree, that is, a Tree with AttributeTreeNodeData and AttributeTreeEdgeData. More... | |
| MassTree | convert_common_tree_to_mass_tree (CommonTree const &source) |
| Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an MassTree, that is, a Tree with MassTreeNodeData and MassTreeEdgeData. More... | |
| SimpleNewickTree | convert_common_tree_to_simple_newick_tree (CommonTree const &source) |
| Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an SimpleNewickTree, that is, a Tree with SimpleNewickNodeData and SimpleNewickEdgeData. More... | |
| CommonTree | convert_to_common_tree (Tree const &source_tree) |
| Convert a Tree to a CommonTree with CommonNodeData and CommonEdgeData. More... | |
| double | deepest_distance (Tree const &tree) |
| Return the longest distance from any point in the tree (on the edges) to any leaf. More... | |
| size_t | degree (TreeLink const &link) |
| Return the degree of the node for a given TreeLink, i.e. how many neighbouring nodes it has. More... | |
| size_t | degree (TreeNode const &node) |
| Return the degree of the node, i.e. how many neighbouring nodes it has. More... | |
| void | delete_edge (Tree &tree, TreeEdge &target_edge, std::function< void(TreeNode &remaining_node, TreeNode &deleted_node)> adjust_nodes={}) |
| Delete a TreeEdge from a Tree, that is, contract the two TreeNodes at its ends into one TreeNode. More... | |
| template<class T > | |
| static void | delete_from_tree_container_ (T &old_elems, std::vector< size_t > &del_elems) |
| Local helper function template that takes one of the tree element containers and deletes all elements at given indices from it. More... | |
| void | delete_leaf_node (Tree &tree, TreeNode &target_node) |
| Delete a leaf TreeNode. More... | |
| void | delete_linear_node (Tree &tree, TreeNode &target_node, std::function< void(TreeEdge &remaining_edge, TreeEdge &deleted_edge)> adjust_edges={}) |
| Delete a "linear" TreeNode from a Tree, that is, a node with two neighbours. More... | |
| void | delete_node (Tree &tree, TreeNode &target_node) |
| Delete a TreeNode from a Tree. More... | |
| void | delete_subtree (Tree &tree, Subtree const &subtree) |
| Delete a complete Subtree from a Tree. More... | |
| void | delete_zero_branch_length_edges (Tree &tree, bool include_leaf_edges=false) |
| Delete (contract) all branches of a CommonTree that have branch length zero. More... | |
| double | diameter (Tree const &tree) |
| Get the diameter of the tree, i.e., the longest distance between any two nodes, measured using the branch_length. More... | |
| double | earth_movers_distance (MassTree const &lhs, MassTree const &rhs, double p=1.0) |
| Calculate the earth mover's distance of two distributions of masses on a given Tree. More... | |
| std::pair< double, double > | earth_movers_distance (MassTree const &tree, double p=1.0) |
| Calculate the earth mover's distance of masses on a given Tree. More... | |
| utils::Matrix< double > | earth_movers_distance (std::vector< MassTree > const &trees, double p=1.0) |
| Calculate the pairwise earth mover's distance for all MassTrees. More... | |
| utils::Matrix< double > | edge_balances (BalanceData const &data, bool reverse_signs=false) |
| Calculate edge balances using the Isometric Log Ratio transformation. More... | |
| TreeEdge * | edge_between (TreeNode &lhs, TreeNode &rhs) |
Return the TreeEdge between two TreeNode&s, if they are neighbours, or nullptr otherwise. More... | |
| TreeEdge const * | edge_between (TreeNode const &lhs, TreeNode const &rhs) |
Return the TreeEdge between two TreeNode&s, if they are neighbours, or nullptr otherwise. More... | |
| utils::Matrix< double > | edge_branch_length_distance_matrix (Tree const &tree) |
| std::vector< double > | edge_branch_length_distance_vector (Tree const &tree, TreeEdge const *edge) |
| std::vector< utils::Color > | edge_color_branch_length_gradient (Tree const &tree, bool zero_based) |
| size_t | edge_count (Tree const &tree) |
| Return the number of Edges of a Tree. Same as Tree::edge_count(). More... | |
| utils::Matrix< size_t > | edge_path_length_matrix (Tree const &tree) |
| std::vector< size_t > | edge_path_length_vector (Tree const &tree, TreeEdge const &edge) |
| utils::Matrix< signed char > | edge_sides (Tree const &tree) |
| Create a Matrix that indiciaces the relative position of the Edges of a Tree, i.e., whether they are on the root side or non-root side. More... | |
| bool | equal (std::vector< Tree > const &trees, std::function< bool(TreeNode const &, TreeNode const &) > node_comparator, std::function< bool(TreeEdge const &, TreeEdge const &) > edge_comparator) |
| Compare all trees for equality given binary comparator functionals for their nodes and edges. More... | |
| bool | equal (Tree const &lhs, Tree const &rhs, std::function< bool(TreeNode const &, TreeNode const &) > node_comparator, std::function< bool(TreeEdge const &, TreeEdge const &) > edge_comparator) |
| Compare two trees for equality given binary comparator functionals for their nodes and edges. More... | |
| bool | equal (TreeSet const &tree_set, std::function< bool(TreeNode const &, TreeNode const &)> node_comparator, std::function< bool(TreeEdge const &, TreeEdge const &)> edge_comparator) |
| Compare whether all Trees in a TreeSet are equal using a given comparator functional. More... | |
| bool | equal_common_trees (Tree const &lhs, Tree const &rhs, bool compare_node_names=true, bool compare_branch_lengths=true) |
| Compare two CommonTrees, that is, check whether they have identical topology, node names, and branch lenghts. More... | |
| template<typename ElementType > | |
| utils::Range< IteratorEulertour< false > > | eulertour (ElementType &element) |
| template<typename ElementType > | |
| utils::Range< IteratorEulertour< true > > | eulertour (ElementType const &element) |
| std::vector< size_t > | find_monophyletic_subtree_edges (Tree const &tree, std::vector< Bipartition > const &bipartitions, std::vector< TreeNode const * > const &nodes, bool include_splitting_edges=true, bool include_leaf_edges=true) |
| Find clades of the tree that are monophyletic with respect to the given list of nodes, that is, clades that only contain nodes from that list. Return all edge indices of those clades. More... | |
| std::vector< size_t > | find_monophyletic_subtree_edges (Tree const &tree, std::vector< std::string > const &node_names, bool include_splitting_edges, bool include_leaf_edges) |
| std::vector< size_t > | find_monophyletic_subtree_edges (Tree const &tree, std::vector< TreeNode const * > const &nodes, bool include_splitting_edges, bool include_leaf_edges) |
| TreeNode * | find_node (Tree &tree, std::string const &name, bool throw_on_failure=false, bool replace_underscores=false) |
| Finds a Node, given its name. If not found, nullptr is returned. More... | |
| TreeNode const * | find_node (Tree const &tree, std::string const &name, bool throw_on_failure=false, bool replace_underscores=false) |
| Finds a Node, given its name. More... | |
| std::vector< TreeNode * > | find_nodes (Tree &tree, std::vector< std::string > const &node_names, bool throw_on_failure=false, bool replace_underscores=false) |
| Find TreeNodes in a Tree, given their name. More... | |
| std::vector< TreeNode const * > | find_nodes (Tree const &tree, std::vector< std::string > const &node_names, bool throw_on_failure=false, bool replace_underscores=false) |
| Find TreeNodes in a Tree, given their name. More... | |
| Bipartition | find_smallest_subtree (Tree const &tree, std::vector< Bipartition > const &bipartitions, std::vector< TreeNode const * > const &nodes) |
| Find the smallest subtree (measured in number of nodes) that contains all given nodes. More... | |
| Tree * | find_tree (TreeSet &tree_set, std::string const &name) |
Get the first Tree in a TreeSet that is stored with a given name, or nullptr if not found. More... | |
| Tree const * | find_tree (TreeSet const &tree_set, std::string const &name) |
Get the first Tree in a TreeSet that is stored with a given name, or nullptr if not found. More... | |
| std::vector< std::pair< TreeNode const *, double > > | furthest_leaf_distance_vector (Tree const &tree) |
| Opposite of closest_leaf_distance_vector(). More... | |
| std::vector< std::pair< TreeNode const *, double > > | furthest_leaf_distance_vector (Tree const &tree, utils::Matrix< double > const &node_distances) |
| std::vector< size_t > | get_clade_edges (Tree const &tree, std::vector< std::string > const &node_names) |
| std::vector< size_t > | get_clade_edges (Tree const &tree, std::vector< tree::TreeNode const * > const &nodes) |
| utils::SvgDocument | get_color_tree_svg_doc_ (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, std::vector< utils::SvgGroup > const &node_shapes, std::vector< utils::SvgGroup > const &edge_shapes) |
| utils::SvgDocument | get_color_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< double > const &value_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm) |
| utils::SvgDocument | get_color_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch) |
| utils::SvgDocument | get_color_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, std::vector< utils::Color > const &color_list, std::vector< std::string > const &color_labels) |
| utils::SvgDocument | get_color_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm) |
| utils::SvgDocument | get_color_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::vector< utils::SvgGroup > const &node_shapes, std::vector< utils::SvgGroup > const &edge_shapes) |
| std::vector< size_t > | get_subtree_edges (TreeLink const &subtree) |
| utils::SvgDocument | get_tree_svg_document (CommonTree const &tree, LayoutParameters const ¶ms) |
| utils::SvgDocument | heat_tree (HeatTreeParameters const ¶ms) |
| utils::SvgDocument | heat_tree (HeatTreeParameters const ¶ms, utils::ColorMap const &matrix_color_map, utils::ColorNormalization const &matrix_color_norm) |
| utils::SvgDocument | heat_tree (HeatTreeParameters const ¶ms, utils::ColorMap const &matrix_color_map, utils::ColorNormalization const &matrix_color_norm, utils::ColorMap const &tree_color_map, utils::ColorNormalization const &tree_color_norm) |
| void | heat_tree_add_heat_matrix_bmp_ (HeatTreeParameters const ¶ms, RectangularLayout const &layout, utils::Matrix< utils::Color > const &matrix, utils::SvgDocument &svg_doc, HeatTreeGrid &grid) |
| void | heat_tree_add_heat_matrix_svg_ (HeatTreeParameters const ¶ms, RectangularLayout const &layout, utils::Matrix< utils::Color > const &matrix, utils::SvgDocument &svg_doc, HeatTreeGrid &grid) |
| void | heat_tree_add_matrix_color_scale_ (utils::ColorMap const &matrix_color_map, utils::ColorNormalization const &matrix_color_norm, utils::SvgDocument &svg_doc, HeatTreeGrid &grid) |
| void | heat_tree_add_tree_color_scale_ (RectangularLayout const &layout, utils::ColorMap const &tree_color_map, utils::ColorNormalization const &tree_color_norm, utils::SvgDocument &svg_doc, HeatTreeGrid &grid) |
| template<class T > | |
| utils::Matrix< T > | heat_tree_reorder_rows_ (utils::Matrix< T > const &mat, std::vector< size_t > const &order) |
| static std::vector< size_t > | heat_tree_row_order_ (Tree const &tree, LayoutSpreading spreading) |
| RectangularLayout | heat_tree_tree_layout_ (HeatTreeParameters const ¶ms) |
| double | height (Tree const &tree) |
| Get the height of the tree, i.e., the longest distance from the root to a leaf, measured using the branch_length. More... | |
| bool | identical_topology (std::vector< Tree > const &trees, bool identical_indices=true) |
| Return whether all trees have an identical topology. More... | |
| bool | identical_topology (Tree const &lhs, Tree const &rhs, bool identical_indices=true) |
| Return whether both trees have an identical topology. More... | |
| bool | identical_topology (TreeSet const &tree_set) |
| Return true iff all Trees in a TreeSet have an identical topology. More... | |
| size_t | inner_edge_count (Tree const &tree) |
| Return the number of Edges of a Tree that do not lead to a leaf Node. More... | |
| std::vector< size_t > | inner_edge_indices (Tree const &tree) |
| Get a list of the edge indices of all inner edges, that is, all TreeEdges that do not lead to a leaf TreeNode. More... | |
| size_t | inner_node_count (Tree const &tree) |
| Count the number of inner Nodes. More... | |
| std::vector< size_t > | inner_node_indices (Tree const &tree) |
| Get a list of the node indices of all inner TreeNodes. More... | |
| bool | is_bifurcating (Tree const &tree, bool loose=false) |
| Return whether the Tree is bifurcating. More... | |
| bool | is_binary (Tree const &tree, bool loose=false) |
| Alias for is_bifurcating(). More... | |
| bool | is_inner (TreeEdge const &edge) |
| Return true iff the secondary node (outwards) of the given edge is an inner node. More... | |
| bool | is_inner (TreeLink const &link) |
| Return true iff the node of the given link is an inner node. More... | |
| bool | is_inner (TreeNode const &node) |
| Return whether the node is an inner node. More... | |
| bool | is_leaf (TreeEdge const &edge) |
| Return true iff the secondary node (outwards) of the given edge is a leaf node. More... | |
| bool | is_leaf (TreeLink const &link) |
| Return true iff the node of the given link is a leaf node. More... | |
| bool | is_leaf (TreeNode const &node) |
| Return whether the node is a leaf/tip. More... | |
| bool | is_root (TreeLink const &link) |
| Return whether the link belongs to the root node of its Tree. More... | |
| bool | is_root (TreeNode const &node) |
| Return whether the node is the root of its Tree. More... | |
| bool | is_rooted (Tree const &tree) |
| Return whether the Tree is rooted, that is, whether the root node has two neighbors. More... | |
| void | ladderize (Tree &tree, LadderizeOrder order=LadderizeOrder::kSmallFirst) |
| Ladderize a Tree, that is, order its subtrees by size. More... | |
| template<class Comparator > | |
| std::vector< std::pair< TreeNode const *, double > > | leaf_distance_vector (Tree const &tree, utils::Matrix< double > const &node_distances, Comparator comp) |
| Local helper function to calculate either closest_leaf_distance_vector() or furthest_leaf_distance_vector(). More... | |
| size_t | leaf_edge_count (Tree const &tree) |
| Return the number of Edges of a Tree that lead to a leaf Node. More... | |
| std::vector< size_t > | leaf_edge_indices (Tree const &tree) |
| Get a list of the edge indices of all leaf edges, that is, all TreeEdges that lead to a leaf TreeNode. More... | |
| utils::Bitvector | leaf_node_bitvector (Tree const &tree, std::vector< TreeNode const * > leaf_nodes) |
Return a Bitvector that has as many entries as the tree has leaf nodes, and is true where the given leaf_nodes are. More... | |
| size_t | leaf_node_count (Tree const &tree) |
| Count the number of leaf Nodes of a Tree. More... | |
| std::vector< size_t > | leaf_node_indices (Tree const &tree) |
| Get a list of the node indices of all leaf TreeNodes. More... | |
| double | length (Tree const &tree) |
| Get the length of the tree, i.e., the sum of all branch lengths. More... | |
| template<typename ElementType > | |
| utils::Range< IteratorLevelorder< false > > | levelorder (ElementType &element) |
| template<typename ElementType > | |
| utils::Range< IteratorLevelorder< true > > | levelorder (ElementType const &element) |
| TreeNode & | lowest_common_ancestor (TreeNode &node_a, TreeNode &node_b) |
| Return the lowest common ancestor of two TreeNodes. More... | |
| TreeNode const & | lowest_common_ancestor (TreeNode const &node_a, TreeNode const &node_b) |
| Return the lowest common ancestor of two TreeNodes. More... | |
| utils::Matrix< size_t > | lowest_common_ancestors (Tree const &tree) |
| Return the lowest common ancestor of each pair of TreeNodes for a given tree, in form of a Matrix of their indices. More... | |
| TreeNode & | make_rooted (Tree &tree, std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> adjust_edges={}) |
| Root a Tree on the first TreeEdge of its current top level TreeNode. More... | |
| TreeNode & | make_rooted (Tree &tree, TreeEdge &target_edge, std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> adjust_edges={}) |
| Root a Tree at a given TreeEdge. More... | |
| Tree | make_tree_from_parents_table (std::vector< std::string > const &child_names, std::vector< std::string > const &parent_names) |
| Create a tree, given lists of child parent pairs. More... | |
| void | make_tree_from_parents_table_add_edge_ (TreeTableHelpers &helpers, std::string const &child_name, std::string const &parent_name, Tree &tree) |
| void | make_unrooted (Tree &tree, std::function< void(TreeEdge &remaining_edge, TreeEdge &deleted_edge)> adjust_edges={}) |
| Unroot a Tree. More... | |
| std::vector< double > | mass_balance (BalanceData const &data, std::unordered_set< size_t > const &numerator_edge_indices, std::unordered_set< size_t > const &denominator_edge_indices) |
| Calculate the balance of edge masses between two sets of edges. More... | |
| double | mass_balance (BalanceData const &data, std::unordered_set< size_t > const &numerator_edge_indices, std::unordered_set< size_t > const &denominator_edge_indices, size_t tree_index) |
| Calculate the balance of edge masses between two sets of edges. More... | |
| BalanceData | mass_balance_data (MassTree const &tree, BalanceSettings settings={}) |
| Calculate the data needed to calculate balances of a MassTree. More... | |
| BalanceData | mass_balance_data (std::vector< MassTree > const &trees, BalanceSettings settings={}) |
| Calculate the data needed to calculate balances on MassTrees. More... | |
| double | mass_tree_binify_masses (MassTree &tree, size_t number_of_bins) |
| Accumulate all masses of a MassTree into bins on each branch. More... | |
| double | mass_tree_center_masses_on_branches (MassTree &tree) |
| Accumulate all masses of a MassTree on the centers of their edges. More... | |
| double | mass_tree_center_masses_on_branches_averaged (MassTree &tree) |
| Accumulate all masses of a MassTree at the average mass position per edge. More... | |
| void | mass_tree_clear_masses (MassTree &tree) |
| Clear all masses of a MassTree, while keeping its topology. More... | |
| std::vector< double > | mass_tree_mass_per_edge (MassTree const &tree) |
Return a std::vector that contains the total mass for each edge of the given MassTree. More... | |
| utils::Matrix< double > | mass_tree_mass_per_edge (std::vector< MassTree > const &mass_trees) |
| Return the total mass for each edge of the given MassTrees. More... | |
| std::vector< std::pair< double, double > > | mass_tree_mass_per_edge_averaged (MassTree const &tree) |
Return a std::vector that contains the total Mass for each edge of the given MassTree (second value), as well as the average position of that mass on the branch (in branch lengths units, first value). More... | |
| MassTree | mass_tree_merge_trees (MassTree const &lhs, MassTree const &rhs, double const scaler_lhs=1.0, double const scaler_rhs=1.0) |
| Merge all masses of two MassTrees into one and return it. More... | |
| void | mass_tree_merge_trees_inplace (MassTree &lhs, MassTree const &rhs, double const scaler_lhs=1.0, double const scaler_rhs=1.0) |
| Merge all masses of two MassTrees by adding them to the first MassTree. More... | |
| void | mass_tree_normalize_masses (MassTree &tree) |
Scale all masses of a MassTree so that they sum up to 1.0. More... | |
| void | mass_tree_reverse_signs (MassTree &tree) |
| Reverse the sign of each mass point on a MassTree. More... | |
| void | mass_tree_scale_masses (MassTree &tree, double factor) |
Scale all masses of a MassTree with the multiplicative factor factor. More... | |
| double | mass_tree_sum_of_masses (MassTree const &tree) |
| Return the total sum of all masses on the MassTree. More... | |
| void | mass_tree_transform_to_unit_branch_lengths (MassTree &tree) |
Set all branch lengths of a MassTree to 1.0, while keeping the relative position of all masses on the branches. More... | |
| bool | mass_tree_validate (MassTree const &tree, double valid_total_mass_difference=-1.0) |
| Validate the data on a MassTree. More... | |
| void | mass_trees_make_average_branch_lengths (std::vector< MassTree > &mass_trees) |
| Change the branch lengths of all trees to their average, and move the masses accordingly in a proportional way. More... | |
| size_t | max_degree (Tree const &tree) |
| Return the highest degree of the Nodes of a Tree. More... | |
| template<class NodeDataType = CommonNodeData, class EdgeDataType = CommonEdgeData> | |
| Tree | minimal_tree () |
| Create a minimal Tree that can be used with manipulation functions such as add_new_node() or add_new_leaf_node() to build a custom tree, including default data types at nodees and edges. More... | |
| Tree | minimal_tree_topology () |
| Create a minimal Tree that can be used with manipulation functions such as add_new_node() or add_new_leaf_node() to build a custom tree. More... | |
| utils::Matrix< double > | node_branch_length_distance_matrix (Tree const &tree) |
| Return a distance matrix containing pairwise distances between all Nodes, using the branch_length of the Edges as distance measurement. More... | |
| std::vector< double > | node_branch_length_distance_vector (Tree const &tree, TreeNode const *node=nullptr) |
| Return a vector containing the distance of all nodes with respect to the given start node, where distance is measured in the sum of branch lengths between the nodes. More... | |
| size_t | node_count (Tree const &tree) |
| Return the number of Nodes of a Tree. Same as Tree::node_count(). More... | |
| template<typename ElementType > | |
| utils::Range< IteratorNodeLinks< false > > | node_links (ElementType &element) |
| template<typename ElementType > | |
| utils::Range< IteratorNodeLinks< true > > | node_links (ElementType const &element) |
| std::vector< std::string > | node_names (std::vector< Tree > const &tree_set, bool leaves_only=false) |
| Returns a set of all TreeNode names of a vector of Trees or a TreeSet. More... | |
| std::vector< std::string > | node_names (Tree const &tree, bool leaves_only=false) |
| Returns a list of all TreeNode names of a Tree. More... | |
| utils::Matrix< size_t > | node_path_length_matrix (Tree const &tree) |
| Return a matrix containing the pairwise depth of all nodes of the tree. More... | |
| std::vector< size_t > | node_path_length_vector (Tree const &tree) |
| Return a vector containing the depth of all nodes with respect to the root node. More... | |
| std::vector< size_t > | node_path_length_vector (Tree const &tree, TreeNode const &node) |
| Return a vector containing the depth of all nodes with respect to the given start node. More... | |
| utils::Matrix< signed char > | node_root_direction_matrix (Tree const &tree) |
| Calculate a Matrix that indicates the nodes on the root side of a given node. More... | |
| std::vector< size_t > | node_to_leaf_map (Tree const &tree) |
| template<typename ElementType > | |
| utils::Range< IteratorPath< false > > | path (ElementType &start, ElementType &finish) |
| template<typename ElementType > | |
| utils::Range< IteratorPath< true > > | path (ElementType const &start, ElementType const &finish) |
| template<typename ElementType > | |
| utils::Range< IteratorPathSet< false > > | path_set (ElementType &start, ElementType &finish, ElementType &lca) |
| template<typename ElementType > | |
| utils::Range< IteratorPathSet< true > > | path_set (ElementType const &start, ElementType const &finish, ElementType const &lca) |
| std::vector< TreeLink const * > | path_to_root (TreeNode const &node) |
| Helper function that finds all TreeLinks between a given TreeNode and the root of the Tree. More... | |
| std::vector< utils::Color > | phylo_factor_clade_colors (Tree const &tree, std::vector< PhyloFactor > const &factors, size_t num_factors=0, PhyloFactorCladeColors colors={}) |
| Return a color for each edge, indicating which factor (phylogenetic unit, clade) it belongs to. More... | |
| std::vector< size_t > | phylo_factor_edge_indices (std::vector< PhyloFactor > const &factors, size_t max_factor=std::numeric_limits< std::size_t >::max()) |
| Get a list of all edges that have factored out by phylogenetic_factorization(). More... | |
| PhyloFactor | phylo_factor_find_best_edge (size_t iteration, BalanceData const &data, std::unordered_set< size_t > const &candidate_edges, PhyloFactorObjectiveFunction objective) |
| Helper function for phylogenetic_factorization() that tries all candidate edges to find the one that maximizes the objective function. More... | |
| std::vector< utils::Color > | phylo_factor_single_factor_colors (Tree const &tree, std::vector< PhyloFactor > const &factors, size_t factor_index, PhyloFactorSingleColors colors={}) |
| Return a color for each edge indicating its role in a single phylogenetic factor. More... | |
| std::unordered_set< size_t > | phylo_factor_subtree_indices (Subtree const &subtree, std::unordered_set< size_t > const &candidate_edges) |
| Helper function for phylogenetic_factorization() to find the constrained subtrees that are split by an edge. More... | |
| std::vector< PhyloFactor > | phylogenetic_factorization (BalanceData const &data, PhyloFactorObjectiveFunction objective, size_t max_iterations=0, std::function< void(size_t iteration, size_t max_iterations)> log_progress={}) |
| Calculate the Phylogenetic Factorization (PhyloFactor) of a set of MassTrees. More... | |
| std::vector< PhyloFactor > | phylogenetic_factorization (BalanceData const &data, std::function< double(std::vector< double > const &balances)> objective, size_t max_iterations=0, std::function< void(size_t iteration, size_t max_iterations)> log_progress={}) |
| Calculate the Phylogenetic Factorization (PhyloFactor) of a set of MassTrees. More... | |
| utils::Matrix< double > | phylogenetic_ilr_transform (BalanceData const &data, bool reverse_signs=false) |
| Calculate the Phylogenetic Isometric Log Ratio transformation. More... | |
| template<typename ElementType > | |
| utils::Range< IteratorPostorder< false > > | postorder (ElementType &element) |
| template<typename ElementType > | |
| utils::Range< IteratorPostorder< true > > | postorder (ElementType const &element) |
| template<typename ElementType > | |
| utils::Range< IteratorPreorder< false > > | preorder (ElementType &element) |
| template<typename ElementType > | |
| utils::Range< IteratorPreorder< true > > | preorder (ElementType const &element) |
| std::string | print_gist (Tree const &tree, long items) |
| std::string | print_info (Tree const &tree) |
| std::string | print_info (TreeEdge const &edge) |
| std::string | print_info (TreeLink const &link) |
| std::string | print_info (TreeNode const &node) |
| size_t | rf_distance_absolute (Tree const &lhs, Tree const &rhs) |
| Compute the absolute RF (Robinson-Foulds) distance metric between two Trees. More... | |
| std::vector< size_t > | rf_distance_absolute (Tree const &lhs, TreeSet const &rhs) |
Compute the absolute RF (Robinson-Foulds) distance metric between a given lhs Tree and all of the trees in the rhs TreeSet. More... | |
| utils::Matrix< size_t > | rf_distance_absolute (TreeSet const &trees) |
Compute the pairwise absolute RF (Robinson-Foulds) distance metric between a set of trees. More... | |
| double | rf_distance_relative (Tree const &lhs, Tree const &rhs) |
| Compute the relative RF (Robinson-Foulds) distance metric between two Trees. More... | |
| std::vector< double > | rf_distance_relative (Tree const &lhs, TreeSet const &rhs) |
Compute the relative RF (Robinson-Foulds) distance metric between a given lhs Tree and all of the trees in the rhs TreeSet. More... | |
| utils::Matrix< double > | rf_distance_relative (TreeSet const &trees) |
Compute the pairwise relative RF (Robinson-Foulds) distance metric between a set of trees. More... | |
| std::vector< utils::Bitvector > | rf_get_bitvectors (Tree const &tree, std::unordered_map< std::string, size_t > const &names) |
| Get all split Bitvectors for a given Tree. More... | |
| template<typename BitvectorProcessor > | |
| void | rf_get_bitvectors_template (Tree const &tree, std::unordered_map< std::string, size_t > const &names, BitvectorProcessor const &process_bitvector) |
| Local helper function template that constructs all bitvectors for the splits of a tree, but allows to customize what to do with them once constructed. More... | |
| std::unordered_map< utils::Bitvector, utils::Bitvector > | rf_get_occurrences (Tree const &lhs, Tree const &rhs) |
| Get an occurrence map for each split found in two trees. More... | |
| std::unordered_map< utils::Bitvector, utils::Bitvector > | rf_get_occurrences (Tree const &lhs, TreeSet const &rhs) |
| Get an occurrence map for each split found in some Trees. More... | |
| std::unordered_map< utils::Bitvector, utils::Bitvector > | rf_get_occurrences (TreeSet const &trees) |
| Get an occurrence map for each split found in the given TreeSet. More... | |
| std::unordered_map< std::string, size_t > | rf_taxon_name_map (Tree const &tree) |
| Get a mapping from taxon names to unique IDs. More... | |
| void | scale_all_branch_lengths (Tree &tree, double factor=1.0) |
| Scale all branch lengths of a Tree by a given factor. More... | |
| void | set_all_branch_lengths (Tree &tree, double length=1.0) |
| Set all branch lengths of a Tree to a given value. More... | |
| utils::Matrix< signed char > | sign_matrix (Tree const &tree, bool compressed=false) |
| Compute the sign matrix or Sequential Binary Partition (SBP) of a Tree. More... | |
| size_t | subtree_max_path_height (Tree const &tree, TreeLink const &link) |
| Calculate the height of a subtree, that is, the maximum path length to a leaf of that subtree, measured in edges between the link and the leaf. More... | |
| std::vector< size_t > | subtree_max_path_heights (Tree const &tree) |
| std::vector< size_t > | subtree_max_path_heights (Tree const &tree, TreeNode const &node) |
| size_t | subtree_size (Tree const &tree, TreeLink const &link) |
| Return the size of the subtree defined by the given TreeLink, measured in number of nodes. More... | |
| std::vector< size_t > | subtree_sizes (Tree const &tree) |
| Calculate the sizes of all subtrees as seen from the root of the tree. More... | |
| std::vector< size_t > | subtree_sizes (Tree const &tree, TreeNode const &node) |
| Calculate the sizes of all subtrees as seen from the given TreeNode. More... | |
| template<class NodeDataType , class EdgeDataType > | |
| bool | tree_data_is (Tree const &tree, bool allow_null=false) |
| Check whether the data of the nodes and edges of the Tree are exactly of the specified data types. More... | |
| template<class NodeDataType , class EdgeDataType > | |
| bool | tree_data_is_derived_from (Tree const &tree, bool allow_null=false) |
| Check whether the data of the nodes and edges of the Tree are derived from the specified data types. More... | |
| bool | validate_topology (Tree const &tree) |
| Validate that all internal pointers of the Tree elements (TreeLinks, TreeNodes, TreeEdges) to each other are correct and that some other invariants are met. More... | |
| void | write_color_tree_to_nexus_file (CommonTree const &tree, std::vector< double > const &value_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::string const &nexus_filename) |
| void | write_color_tree_to_nexus_file (CommonTree const &tree, std::vector< utils::Color > const &color_per_branch, std::string const &nexus_filename) |
| void | write_color_tree_to_phyloxml_file (CommonTree const &tree, std::vector< double > const &value_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::string const &phyloxml_filename) |
| void | write_color_tree_to_phyloxml_file (CommonTree const &tree, std::vector< utils::Color > const &color_per_branch, std::string const &phyloxml_filename) |
| void | write_color_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< double > const &value_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::string const &svg_filename) |
| void | write_color_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, std::string const &svg_filename) |
| void | write_color_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, std::vector< utils::Color > const &color_list, std::vector< std::string > const &color_labels, std::string const &svg_filename) |
| void | write_color_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::string const &svg_filename) |
| void | write_color_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::vector< utils::Color > const &color_per_branch, utils::ColorMap const &color_map, utils::ColorNormalization const &color_norm, std::vector< utils::SvgGroup > const &node_shapes, std::vector< utils::SvgGroup > const &edge_shapes, std::string const &svg_filename) |
| void | write_tree_to_newick_file (CommonTree const &tree, std::string const &newick_filename) |
| Write a newick file containing a tree. More... | |
| void | write_tree_to_nexus_file (CommonTree const &tree, std::string const &nexus_filename) |
| Write a nexus file containing a tree. More... | |
| void | write_tree_to_phyloxml_file (CommonTree const &tree, std::string const &phyloxml_filename) |
| Write a phyloxml file containing a tree. More... | |
| void | write_tree_to_svg_file (CommonTree const &tree, LayoutParameters const ¶ms, std::string const &svg_filename) |
Enumerations | |
| enum | LadderizeOrder { kSmallFirst, kLargeFirst } |
| enum | LayoutShape { kCircular, kRectangular } |
| Shape of the tree for drawing, either circular or rectangular. More... | |
| enum | LayoutSpreading { kLeafNodesOnly, kAllNodesButRoot, kAllNodes } |
| Spreading of the nodes of a tree for drawing. More... | |
| enum | LayoutType { kPhylogram, kCladogram } |
| Type of tree for drawing, either phylogram or cladogram. More... | |
Typedefs | |
| using | AttributeTree = Tree |
| Alias for a Tree that stores TreeNodes and TreeEdges with string attributes on them. More... | |
| using | AttributeTreeEdge = TreeEdge |
| Alias for a TreeEdge of an AttributeTree. See there for more information. More... | |
| using | AttributeTreeLink = TreeLink |
| Alias for a TreeLink of an AttributeTree. See there for more information. More... | |
| using | AttributeTreeMap = std::map< std::string, std::string > |
| Alias for the map type used by an AttributeTree. More... | |
| using | AttributeTreeNode = TreeNode |
| Alias for a TreeNode of an AttributeTree. See there for more information. More... | |
| using | CommonTree = Tree |
| Alias for a Tree with data types CommonNodeData and CommonEdgeData. More... | |
| using | CommonTreeEdge = TreeEdge |
| Alias for a TreeEdge of a CommonTree. See there for more information. More... | |
| using | CommonTreeLink = TreeLink |
| Alias for a TreeLink of a CommonTree. See there for more information. More... | |
| using | CommonTreeNode = TreeNode |
| Alias for a TreeNode of a CommonTree. See there for more information. More... | |
| using | LayoutTree = tree::Tree |
| Alias for a tree::Tree used for a tree with information needed for tree drawing. More... | |
| using | LayoutTreeEdge = tree::TreeEdge |
| Alias for tree::TreeEdge used in a LayoutTree. See LayoutEdgeData for the data stored on the edges. More... | |
| using | LayoutTreeLink = tree::TreeLink |
| Alias for tree::TreeLink used in a LayoutTree. More... | |
| using | LayoutTreeNode = tree::TreeNode |
| Alias for tree::TreeNode used in a LayoutTree. See LayoutNodeData for the data stored on the nodes. More... | |
| using | MassTree = Tree |
| Alias for a Tree that stores masses on its TreeEdges. More... | |
| using | MassTreeEdge = TreeEdge |
| Alias for a TreeEdge of a MassTree. See there for more information. More... | |
| using | MassTreeLink = TreeLink |
| Alias for a TreeLink of a MassTree. See there for more information. More... | |
| using | MassTreeNode = TreeNode |
| Alias for a TreeNode of a MassTree. See there for more information. More... | |
| using | PhyloFactorObjectiveFunction = std::function< double(size_t iteration, size_t edge_index, std::vector< double > const &balances) > |
| Function type used as the objective function in phylogenetic_factorization(). More... | |
| using | SimpleNewickTree = Tree |
| Alias for a Tree that stores TreeNodes and TreeEdges with the standard Newick elements on them. More... | |
| using | SimpleNewickTreeEdge = TreeEdge |
| Alias for a TreeEdge of an SimpleNewickTree. See there for more information. More... | |
| using | SimpleNewickTreeLink = TreeLink |
| Alias for a TreeLink of an SimpleNewickTree. See there for more information. More... | |
| using | SimpleNewickTreeNode = TreeNode |
| Alias for a TreeNode of an SimpleNewickTree. See there for more information. More... | |
| TreeNode & add_new_leaf_node | ( | Tree & | tree, |
| TreeEdge & | target_edge, | ||
| std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> | adjust_edges = {} |
||
| ) |
Add a new Node as a leaf to an existing Edge, by also adding a new Node in the middle of that Edge.
This function is a combination of add_new_node( Tree&, TreeNode& ) and add_new_node( Tree&, TreeEdge& ). Before adding the new leaf node, it first adds another Node that splits the given target_edge into two edges, and then adds the leaf to it.
Thus, the procedure is as shown:
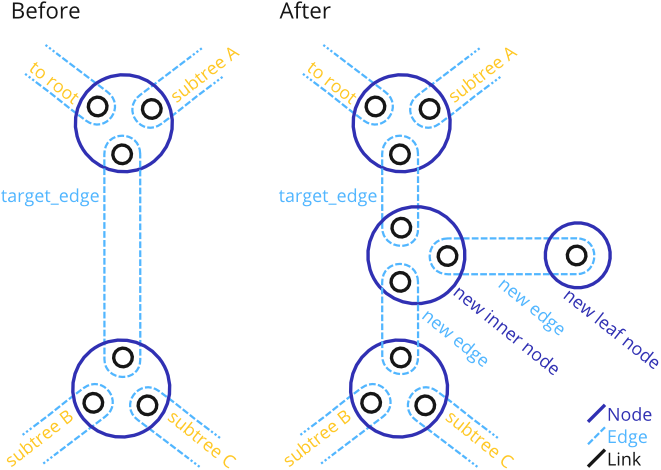
After the function, the target_edge with all its data ends up on the to-root side of the new inner node.
For details of how the data pointers are handled, see add_new_node( Tree&, TreeNode& ). This function behaves in a similar way. The data objects of the new nodes and edges are default-constructed objects of the same type as the target_edge and its primary node.
Be aware that the data of target_edge is not changed by default. Thus, in trees with CommonEdgeData, the branch lengths of the affected edges might have to be changed to the desired values after calling this function. In particular, this concerns the target edge as well as the new inner edge. One can use the adjust_edges functional to adjust these edges. See add_new_node( Tree&, TreeEdge& ) for an example of this.
Definition at line 263 of file tree/function/manipulation.cpp.
| TreeNode & add_new_node | ( | Tree & | tree, |
| TreeEdge & | target_edge, | ||
| std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> | adjust_edges = {} |
||
| ) |
Add a new Node that splits an existing Edge.
This function adds a new Node that splits the given target_edge into two edges. Thus, the procedure is as shown:
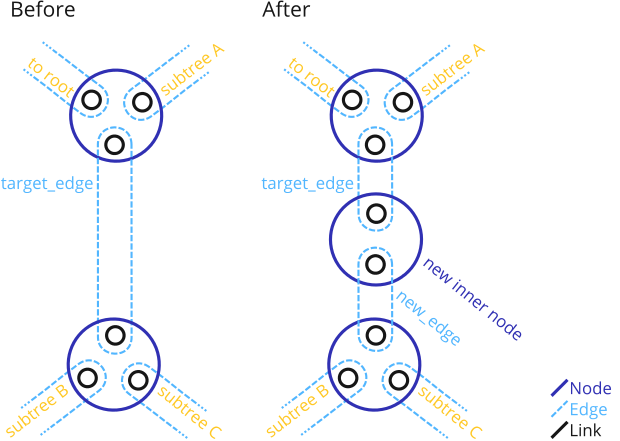
After the function, the target_edge with all its data ends up on the to-root side of the new inner node.
For details of how the data pointers are handled, see add_new_node( Tree&, TreeNode& ). This function behaves in a similar way. The data objects of the new nodes and edges are default-constructed objects of the same type as the target_edge and its primary node.
Be aware that the data of target_edge is not changed by default. Thus, in trees with CommonEdgeData, the branch lengths of the two affected edges might have to be changed to the desired values after calling this function. Instead, one can use the adjust_edges functional to adjust these edges. For example, in order to split the branch length in half between the two edges, use:
// Some tree and edge.
Tree tree;
auto& target_edge = ...;
add_new_node( tree, target_edge, []( TreeEdge& target_edge, TreeEdge& new_edge ){
auto& target_bl = target_edge.data<CommonEdgeData>().branch_length;
auto& new_bl = new_edge.data<CommonEdgeData>().branch_length;
new_bl = target_bl / 2.0;
target_bl = target_bl / 2.0;
});
The functor is called after all changes to the Tree have been made, and all data objects have been created.
Definition at line 188 of file tree/function/manipulation.cpp.
Add a new Node as a leaf to an existing Node.
The function adds a new leaf to the Tree by appending it to an existing TreeNode. For this, four new elements are created and added to the tree:
Thus, the procedure is as shown:
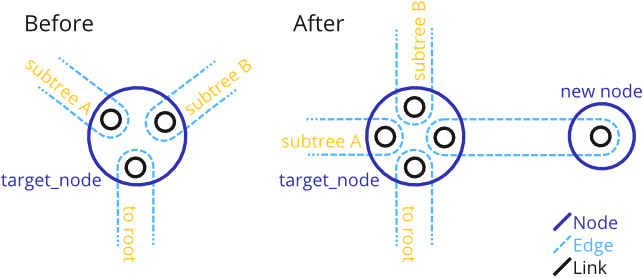
The new Node is added at the "end" of the given Node. That is, when traversing the Tree or the given Node, the new Node will be visited last. In the example, the tree is bifurcating before the new node is added. This is just an example - the function also works with Nodes that are already multifurcating.
The data pointers of the new Node and Edge are initialized to default-constructed objects:
target_node.target_node.This means that in case of a Tree where every Node and Edge has the same node and edge data type (standard case), the newly created ones will also have these data types.
Definition at line 109 of file tree/function/manipulation.cpp.
Return a Tree where the branch lengths are the average of the Trees in the given vector of Trees or TreeSet, given that they all have the same topology.
The function works only under the following conditions:
* All trees must have the same topology. * The TreeType must provide a data member `branch_length` for the edges.
Otherwise, the function throws an std::runtime_error. It does not check for node names, but the returned tree will contain the names of the first tree in the set.
Definition at line 242 of file tree/common_tree/functions.cpp.
Return whether the Subtree belongs to the Tree, i.e., whether its link is owned by the Tree.
Definition at line 255 of file tree/function/operators.cpp.
Return whether the Subtree belongs to the Tree, i.e., whether its link is owned by the Tree.
Definition at line 250 of file tree/function/operators.cpp.
Return whether the TreeEdge belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 230 of file tree/function/operators.cpp.
Return whether the TreeLink belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 240 of file tree/function/operators.cpp.
Return whether the TreeNode belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 220 of file tree/function/operators.cpp.
Return whether the TreeEdge belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 235 of file tree/function/operators.cpp.
Return whether the TreeLink belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 245 of file tree/function/operators.cpp.
Return whether the TreeNode belongs to the Tree, i.e., whether it is owned by the Tree.
Definition at line 225 of file tree/function/operators.cpp.
| std::vector< Bipartition > bipartition_set | ( | Tree const & | tree | ) |
Definition at line 58 of file tree/bipartition/functions.cpp.
| std::vector< double > branch_lengths | ( | Tree const & | tree | ) |
Get a vector of all branch lengths of a Tree, index by the edge index.
Definition at line 213 of file tree/common_tree/functions.cpp.
"Reroot" the Tree at the given TreeLink.
The function sets the root of the tree to the node of the given link. This operation does not change the topology of the tree, but merely adjusts some internal properties. The main changes are that Tree::root_node() and Tree::root_link() will return the new root after calling this function, and that tree iterators will start traversing the tree from this new root by default.
There are three internal changes made to the tree data structure:
The difference between this function and change_rooting( Tree& tree, TreeNode const& ) is that when specifying a specific link, this link is used as the (primary) link of the new root node. This way, algorithms and iterators (e.g., IteratorLevelorder) will start traversing the tree in the direction of this link by default. When specifying a node for rerooting instead, the primary link of that node is used, so that iterators start traversing the tree in the direction of the old root instead. For most applications, this does not make a difference. However, there might be cases where the start directing makes a difference. Thus, we offer both versions of this function.
The link needs to be part of the tree, otherwise an exception is thrown.
Definition at line 779 of file tree/function/manipulation.cpp.
Reroot the Tree at the given TreeNode.
See change_rooting( Tree&, TreeLink& ) for details.
The node needs to be part of the tree, otherwise an exception is thrown.
Definition at line 828 of file tree/function/manipulation.cpp.
Returns a vector containing the closest leaf node for each node, measured in number of edges between them and its depth (number of edges between them).
The vector is indexed using the node().index() for every node. Its value contains an std::pair, where the first element is a NodeType* to the closest leaf node (with respect to its depth) and the second element its depth with respect to the node at the given index of the vector. The depth is the number of edges visited on the path between two nodes (0 for itself, 1 for immediate neighbours, etc).
Thus, leaf nodes will have a pointer to themselves and a depth value of 0, and for all other nodes the depth will be the number of edges between it and the closest leaf node.
There might be more than one leaf with the same depth to a given node. In this case, an arbitrary one is used.
Definition at line 251 of file tree/function/distances.cpp.
| std::vector< std::pair< TreeNode const *, double > > closest_leaf_distance_vector | ( | Tree const & | tree | ) |
Return a vector containing the closest leaf node for each node, using the branch_length as distance measure.
The vector is indexed using the node().index() for every node. Its value contains an std::pair, where the first element is a TreeNode* to the closest leaf node of the node at the index, measured using the branch_length; the second element of the pair is the distance value itself. Thus, leaf nodes will have a pointer to themselves and a distance value of 0.
See also furthest_leaf_distance_vector().
Definition at line 345 of file tree/common_tree/distances.cpp.
| std::vector< std::pair< TreeNode const *, double > > closest_leaf_distance_vector | ( | Tree const & | tree, |
| utils::Matrix< double > const & | node_distances | ||
| ) |
Definition at line 354 of file tree/common_tree/distances.cpp.
| Tree convert | ( | Tree const & | source, |
| std::function< std::unique_ptr< BaseNodeData >(BaseNodeData const &node_data)> | node_data_converter, | ||
| std::function< std::unique_ptr< BaseEdgeData >(BaseEdgeData const &edge_data)> | edge_data_converter | ||
| ) |
Create a tree with the same topology as the source tree, while converting its data.
This function takes the given source Tree (possibly with different data types at the nodes and edges), and copies its topology (i.e., all links, nodes and edges, and their structure) to the newly created result tree.
The data types are then converted using the two provided functions for the node data type and edge data type, respectively. If a node or an edge does not have data (i.e., the data pointer is a nullptr), the converter functions are not called, but the data of the new tree at that node or edge is also set to a nullptr.
Definition at line 53 of file tree/function/operators.cpp.
|
inline |
Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an AttributeTree, that is, a Tree with AttributeTreeNodeData and AttributeTreeEdgeData.
Definition at line 211 of file tree/attribute_tree/tree.hpp.
|
inline |
Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an MassTree, that is, a Tree with MassTreeNodeData and MassTreeEdgeData.
Definition at line 213 of file tree/mass_tree/tree.hpp.
|
inline |
Helper function that takes a CommonTree (or any Tree with Node and Edge data derived from it) and turns its data into an SimpleNewickTree, that is, a Tree with SimpleNewickNodeData and SimpleNewickEdgeData.
Definition at line 229 of file simple_tree.hpp.
| CommonTree convert_to_common_tree | ( | Tree const & | source_tree | ) |
Convert a Tree to a CommonTree with CommonNodeData and CommonEdgeData.
This works for all Trees that have data derived from those two data base classes. Also nodes or edges without data work. However, an expection is thrown if any node or edge contains data that is not derived from CommonNodeData and CommonEdgeData, respectively.
The data itself is copied using the clone functions, so that all names and branch lengths are transferred to the returned Tree.
Definition at line 95 of file tree/common_tree/operators.cpp.
| double deepest_distance | ( | Tree const & | tree | ) |
Return the longest distance from any point in the tree (on the edges) to any leaf.
Definition at line 275 of file tree/common_tree/distances.cpp.
| size_t degree | ( | TreeLink const & | link | ) |
Return the degree of the node for a given TreeLink, i.e. how many neighbouring nodes it has.
Definition at line 103 of file tree/function/functions.cpp.
| size_t degree | ( | TreeNode const & | node | ) |
Return the degree of the node, i.e. how many neighbouring nodes it has.
Definition at line 108 of file tree/function/functions.cpp.
| void delete_edge | ( | Tree & | tree, |
| TreeEdge & | target_edge, | ||
| std::function< void(TreeNode &remaining_node, TreeNode &deleted_node)> | adjust_nodes = {} |
||
| ) |
Delete a TreeEdge from a Tree, that is, contract the two TreeNodes at its ends into one TreeNode.
This can be understood as contracting the tree at an edge, pulling both nodes at the ends of the edge together and merging them into one node with all subtrees of the original nodes combined. The TreeNode that remains is the one closer to the root. The other TreeNode is deleted, and all its outgoing branches are re-attached to the reamining node. If that other (to-be-deleted) TreeNode is a leaf, it is simply fully removed.
Using the adjust_nodes function, the properties of these two TreeNodes can be adjusted before deletion, for example transferring some data from the deleted node to the remaining node.
Definition at line 597 of file tree/function/manipulation.cpp.
|
static |
Local helper function template that takes one of the tree element containers and deletes all elements at given indices from it.
The function modifies both its parameters: The deletion list gets sorted.
Definition at line 476 of file tree/function/manipulation.cpp.
Delete a leaf TreeNode.
If the deleted node is the root, the root is reset to the node where the leaf is attached.
Definition at line 296 of file tree/function/manipulation.cpp.
| void delete_linear_node | ( | Tree & | tree, |
| TreeNode & | target_node, | ||
| std::function< void(TreeEdge &remaining_edge, TreeEdge &deleted_edge)> | adjust_edges = {} |
||
| ) |
Delete a "linear" TreeNode from a Tree, that is, a node with two neighbours.
Such nodes for example occur as the root node in properly rooted trees. The deletion of this node leads to its two edges becoming one. The edge that is deleted is the one further away from the root of the tree, while the other edge remains. If the target_node itself is the root, the edge that is being deleted is the one that is not at the primary link of the node. In that case, the root is also reset to the node adjacent to the primary link of the target_node.
As one edge is deleted, it might be neccessary to update the data of the other, for example, to adjust its branch length by adding up both of them. The adjust_edges functional can be used to achivve this. By default, no adjustments are done. For an example of a similar function, see add_new_node( Tree&, TreeEdge& ).
Definition at line 385 of file tree/function/manipulation.cpp.
Delete a TreeNode from a Tree.
This function is a simple wrapper for the specialized deletion functions:
target_node away from the root.See these other functions for details.
Definition at line 277 of file tree/function/manipulation.cpp.
Delete a complete Subtree from a Tree.
The function deletes a Subtree, including the edge where it is attached. This works for almost all possible subtrees (including leaf-only subtrees), with one notable exception: A subtree that contains all of the tree but one leaf cannot be deleted. In that case, the remaining tree would be just a leaf, with no link and no edge, which is not a valid tree.
If the subtree contains the root node, the root is reset to the node where the subtree is attached.
Definition at line 518 of file tree/function/manipulation.cpp.
| void delete_zero_branch_length_edges | ( | Tree & | tree, |
| bool | include_leaf_edges = false |
||
| ) |
Delete (contract) all branches of a CommonTree that have branch length zero.
See delete_edge() for details on the underlying function. We here additonally adjust node names so that leaf node names are transferred to remaining inner nodes as far as possible.
By default, the function does not delete edges that lead to leaf nodes, even if those edges have a branch length of 0. If include_leaf_edges is set to true however, these edges are deleted as well, and their leaf node names (if present) are transferred to their parent node if possible. As a node can only have one name, that means that nodes that have multiple attached leaf nodes will only get one of the leaf node names (depending on the ordering of edge indices), while the other leaf node names will be deleted with their respetive nodes.
Definition at line 706 of file tree/function/manipulation.cpp.
| double diameter | ( | Tree const & | tree | ) |
Get the diameter of the tree, i.e., the longest distance between any two nodes, measured using the branch_length.
Definition at line 178 of file tree/common_tree/functions.cpp.
Calculate the earth mover's distance of two distributions of masses on a given Tree.
The earth mover's distance is typically a distance measure between two distributions. See [Earth mover's distance](https://en.wikipedia.org/wiki/Earth_mover's_distance) for an introduction.
In our case, we use distibutions of masses along the branches of a tree. Each branch can have multiple masses at different positions within [0.0, branch_length].
The distance is calculated as the amount of work needed to move the masses of one distribution so that they end up in the positions of the masses of the other distribution. Work is here defined as mass times dislocation. Thus, the work is higher if either more mass has to be moved, or if mass has to be moved further.
Here, the parameter p is used to control the influence of mass and distance, with 0.0 < p < inf, and default p == 1.0, which is the neutral case. A larger p increases the impact of distance traveled, while a smaller p emphasizes differences of mass. For details, see the references cited below.
The resulting distance is independed of the rooting of the tree and commutative with respect to the two mass distributions.
The earth mover's distance is only meaningful if both mass distributions contain the same amount of total mass. See mass_tree_sum_of_masses() to check this. Also, in order to give comparable results over different tree topologies, the mass can be normalized using mass_tree_normalize_masses(). Then, the result of the earth mover's distance is always in the range [ 0.0, 1.0 ].
See earth_movers_distance( Sample const&, ... ) for an exemplary usage of this function, which applies the earth mover's distance to the placement weights (like_weight_ratio) of a PlacementTree.
References:
[1] Guppy Documentation: http://matsen.github.io/pplacer/generated_rst/guppy_kr.html#guppy-kr
[2] F. A. Matsen and S. N. Evans, “Edge principal components and squash clustering: using the special structure of phylogenetic placement data for sample comparison.”, PLoS One, 2011. DOI: 10.1371/journal.pone.0056859
[3] S. N. Evans and F. A. Matsen, “The phylogenetic Kantorovich-Rubinstein metric for environmental sequence samples.”, Statistical Methodology, 2012. DOI: 10.1111/j.1467-9868.2011.01018.x
Definition at line 60 of file tree/mass_tree/emd.cpp.
| std::pair< double, double > earth_movers_distance | ( | MassTree const & | tree, |
| double | p = 1.0 |
||
| ) |
Calculate the earth mover's distance of masses on a given Tree.
This function is mainly used as a speed-up for calculating earth_movers_distance( MassTree const&, MassTree const& ). See there for more details.
It uses the following convention for the two distributions: The masses of one distribution are stored using a positive sign, the masses of the other distribution use a negative sign. This way, only one Tree needs to be stored, and the algorithm is significantly simplyfied.
Thus, as the earth mover's distance is only meaningful if both distributions have the same sum, and we use opposite signs to store the masses, the sum of all masses on the tree should ideally be zero (apart from numerical derivations). See mass_tree_sum_of_masses() and mass_tree_validate() for functions to verify this.
The function returns two doubles: The first one is the actual distance, the second one gives the remaining mass at the root node. This should also be close to 0.0, as there, all masses from the subtrees should ideally cancel each other out. Use this value to check whether this actually worked out. Too big numbers indicate that something is wrong with the sums of the signed masses.
Definition at line 237 of file tree/mass_tree/emd.cpp.
| utils::Matrix< double > earth_movers_distance | ( | std::vector< MassTree > const & | trees, |
| double | p = 1.0 |
||
| ) |
Calculate the pairwise earth mover's distance for all MassTrees.
The result is a pairwise distance Matrix using the indices of the given vector. See earth_movers_distance( MassTree const&, MassTree const&, double ) for details on the calculation.
Definition at line 183 of file tree/mass_tree/emd.cpp.
| utils::Matrix< double > edge_balances | ( | BalanceData const & | data, |
| bool | reverse_signs = false |
||
| ) |
Calculate edge balances using the Isometric Log Ratio transformation.
This is a hybrid method between the phylogenetic_ilr_transform() and edge imbalances: We calcualte the balance between the masses on the two sides of the split induced by each edge. This is similar to edge imbalances, in that it splits the tree at each edge, but instead of calculating the imbalance, we use the ILR transform to calculate balances instead.
Definition at line 153 of file phylo_ilr.cpp.
Return the TreeEdge between two TreeNode&s, if they are neighbours, or nullptr otherwise.
Definition at line 260 of file tree/function/operators.cpp.
Return the TreeEdge between two TreeNode&s, if they are neighbours, or nullptr otherwise.
Definition at line 272 of file tree/function/operators.cpp.
| utils::Matrix< double > edge_branch_length_distance_matrix | ( | Tree const & | tree | ) |
Definition at line 155 of file tree/common_tree/distances.cpp.
| std::vector< double > edge_branch_length_distance_vector | ( | Tree const & | tree, |
| TreeEdge const * | edge | ||
| ) |
Definition at line 221 of file tree/common_tree/distances.cpp.
| std::vector< utils::Color > edge_color_branch_length_gradient | ( | Tree const & | tree, |
| bool | zero_based | ||
| ) |
Definition at line 51 of file tree/common_tree/edge_color.cpp.
| size_t edge_count | ( | Tree const & | tree | ) |
Return the number of Edges of a Tree. Same as Tree::edge_count().
Definition at line 212 of file tree/function/functions.cpp.
| utils::Matrix< size_t > edge_path_length_matrix | ( | Tree const & | tree | ) |
Definition at line 144 of file tree/function/distances.cpp.
Definition at line 202 of file tree/function/distances.cpp.
| utils::Matrix< signed char > edge_sides | ( | Tree const & | tree | ) |
Create a Matrix that indiciaces the relative position of the Edges of a Tree, i.e., whether they are on the root side or non-root side.
For a Tree with e many Edges, the resulting square Matrix has dimensions ( e, e ). The row at index i corresponds to edge i, which can be accessed via Tree::edge_at(). The entries of that row indicate whether the other edges (as indexed by the columns) are on the root side of the edge i, in which case the entry is -1, or on the non-root side, in which case the entry is 1. The entry for the edge i itself (which is the diagonale) is 0.
In other words, all edges on the proximal side of a given edge are denoted by a -1, while all edges on the distal side of the given edge are denoted by a 1.
As a consequence, the rows of edges that lead to a leaf are full of 1s (with the exception of the diagonal entry, which still is 0). This is because for leaf edges, all other edges are on the root side of the tree.
Definition at line 265 of file tree/function/functions.cpp.
| bool equal | ( | std::vector< Tree > const & | trees, |
| std::function< bool(TreeNode const &, TreeNode const &) > | node_comparator, | ||
| std::function< bool(TreeEdge const &, TreeEdge const &) > | edge_comparator | ||
| ) |
Compare all trees for equality given binary comparator functionals for their nodes and edges.
Definition at line 118 of file tree/function/operators.cpp.
| bool equal | ( | Tree const & | lhs, |
| Tree const & | rhs, | ||
| std::function< bool(TreeNode const &, TreeNode const &) > | node_comparator, | ||
| std::function< bool(TreeEdge const &, TreeEdge const &) > | edge_comparator | ||
| ) |
Compare two trees for equality given binary comparator functionals for their nodes and edges.
This function does a preorder traversal of both trees in parallel and calls the comparator functionals for each position of the iterator. It returns true iff the comparators are true for every position.
The comparator functionals can be either function pointers, function objects, or lambda expressions.
As the traversal is done in parallel, the trees are also checked for equal topology: their elements (links, nodes, edges) have to be equal in size and the degree of each node during the traversal has to be identical in both trees. Those assumptions are made because two trees that do not have identical topology are never considered equal.
Definition at line 80 of file tree/function/operators.cpp.
| bool equal | ( | TreeSet const & | tree_set, |
| std::function< bool(TreeNode const &, TreeNode const &)> | node_comparator, | ||
| std::function< bool(TreeEdge const &, TreeEdge const &)> | edge_comparator | ||
| ) |
Compare whether all Trees in a TreeSet are equal using a given comparator functional.
See equal() for more information.
Definition at line 72 of file tree_set.cpp.
| bool equal_common_trees | ( | Tree const & | lhs, |
| Tree const & | rhs, | ||
| bool | compare_node_names, | ||
| bool | compare_branch_lengths | ||
| ) |
Compare two CommonTrees, that is, check whether they have identical topology, node names, and branch lenghts.
Definition at line 46 of file tree/common_tree/operators.cpp.
| utils::Range< IteratorEulertour< false > > genesis::tree::eulertour | ( | ElementType & | element | ) |
Definition at line 216 of file eulertour.hpp.
| utils::Range< IteratorEulertour< true > > genesis::tree::eulertour | ( | ElementType const & | element | ) |
Definition at line 206 of file eulertour.hpp.
| std::vector< size_t > find_monophyletic_subtree_edges | ( | Tree const & | tree, |
| std::vector< Bipartition > const & | bipartitions, | ||
| std::vector< TreeNode const * > const & | nodes, | ||
| bool | include_splitting_edges = true, |
||
| bool | include_leaf_edges = true |
||
| ) |
Find clades of the tree that are monophyletic with respect to the given list of nodes, that is, clades that only contain nodes from that list. Return all edge indices of those clades.
The function takes a set of leaf nodes, and selects all branches of the tree that belong to monophyletic clades containing only leaf nodes from the given set:
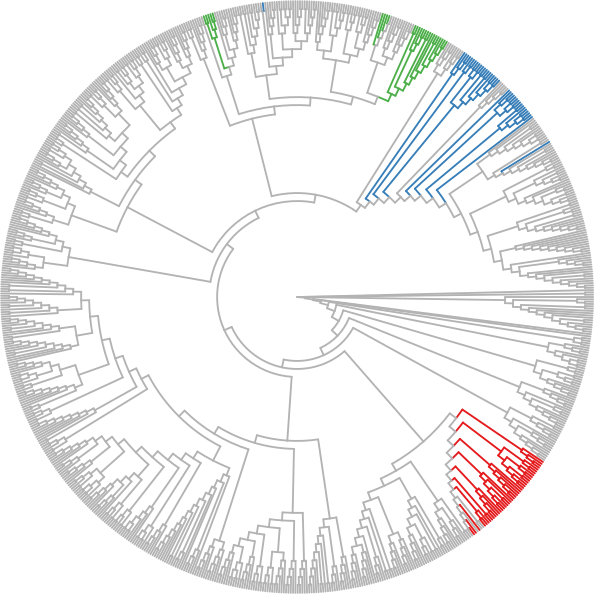
In the example, the function was called three times with three different sets of nodes. In each case, all branches are selected/colorized that are monophyletic with respect to the nodes. In other words, the function conceptually iterates all edges of the tree. If one side of the split induced by an edge only contains leaf nodes from the given set, the whole clade is monophyletic with respect to that set, and hence added to the resulting list of edges.
If include_splitting_edge is true (default), the edges that separate each clade from the rest of the tree are also included. This is particularly important for edges leading to a leaf/tip of the tree: If set to false, those edges are not included, meaning that the respective node does not contribute to the result at all.
In order to solve/refine this, that is to not include the splitting edge of larger clades, but still include an edge that leads to a single leaf node (if this node is not part of any larger clade), the additional parameter include_leaf_edges can be used. It also defaults to true, meaning that those edges are included by default.
Definition at line 275 of file tree/bipartition/functions.cpp.
| std::vector< size_t > find_monophyletic_subtree_edges | ( | Tree const & | tree, |
| std::vector< std::string > const & | node_names, | ||
| bool | include_splitting_edges, | ||
| bool | include_leaf_edges | ||
| ) |
Definition at line 354 of file tree/bipartition/functions.cpp.
| std::vector< size_t > find_monophyletic_subtree_edges | ( | Tree const & | tree, |
| std::vector< TreeNode const * > const & | nodes, | ||
| bool | include_splitting_edges, | ||
| bool | include_leaf_edges | ||
| ) |
Definition at line 342 of file tree/bipartition/functions.cpp.
| TreeNode * find_node | ( | Tree & | tree, |
| std::string const & | name, | ||
| bool | throw_on_failure = false, |
||
| bool | replace_underscores = false |
||
| ) |
Finds a Node, given its name. If not found, nullptr is returned.
Finds a Node, given its name. If not found, by default, nullptr is returned. If however throw_on_failure is set to true, an exception is thrown instead. This is useful if the continuation of the calling function does not make sense without having found the node.
Definition at line 109 of file tree/common_tree/functions.cpp.
| TreeNode const * find_node | ( | Tree const & | tree, |
| std::string const & | name, | ||
| bool | throw_on_failure = false, |
||
| bool | replace_underscores = false |
||
| ) |
Finds a Node, given its name.
If not found, by default, nullptr is returned. If however throw_on_failure is set to true, an exception is thrown instead. This is useful if the continuation of the calling function does not make sense without having found the node.
Definition at line 84 of file tree/common_tree/functions.cpp.
| std::vector< TreeNode * > find_nodes | ( | Tree & | tree, |
| std::vector< std::string > const & | node_names, | ||
| bool | throw_on_failure = false, |
||
| bool | replace_underscores = false |
||
| ) |
Find TreeNodes in a Tree, given their name.
Find TreeNodes in a Tree, given their name. If a particular node is not found, by default, the respective entry is a nullptr. If however throw_on_failure is set to true, an exception is thrown instead. This is useful if the continuation of the calling function does not make sense without having found the node.
Definition at line 139 of file tree/common_tree/functions.cpp.
| std::vector< TreeNode const * > find_nodes | ( | Tree const & | tree, |
| std::vector< std::string > const & | node_names, | ||
| bool | throw_on_failure = false, |
||
| bool | replace_underscores = false |
||
| ) |
Find TreeNodes in a Tree, given their name.
If a particular node is not found, by default, the respective entry is a nullptr. If however throw_on_failure is set to true, an exception is thrown instead. This is useful if the continuation of the calling function does not make sense without having found the node.
Definition at line 122 of file tree/common_tree/functions.cpp.
| Bipartition find_smallest_subtree | ( | Tree const & | tree, |
| std::vector< Bipartition > const & | bipartitions, | ||
| std::vector< TreeNode const * > const & | nodes | ||
| ) |
Find the smallest subtree (measured in number of nodes) that contains all given nodes.
A subtree is defined by one of the two parts of a tree that are splitted by one edge. Thus, conceptually, this function tries all subtrees by leaving out each edge once. It then returns the smalles subtree that contains all of the given nodes.
The subtree might contain additional nodes that are not in the given set. If no fitting subtree exists, the function returns an empty Bipartition.
Definition at line 215 of file tree/bipartition/functions.cpp.
Get the first Tree in a TreeSet that is stored with a given name, or nullptr if not found.
Definition at line 48 of file tree_set.cpp.
Get the first Tree in a TreeSet that is stored with a given name, or nullptr if not found.
Definition at line 58 of file tree_set.cpp.
| std::vector< std::pair< TreeNode const *, double > > furthest_leaf_distance_vector | ( | Tree const & | tree | ) |
Opposite of closest_leaf_distance_vector().
Definition at line 361 of file tree/common_tree/distances.cpp.
| std::vector< std::pair< TreeNode const *, double > > furthest_leaf_distance_vector | ( | Tree const & | tree, |
| utils::Matrix< double > const & | node_distances | ||
| ) |
Definition at line 370 of file tree/common_tree/distances.cpp.
| std::vector< size_t > get_clade_edges | ( | Tree const & | tree, |
| std::vector< std::string > const & | node_names | ||
| ) |
Definition at line 382 of file tree/bipartition/functions.cpp.
| std::vector< size_t > get_clade_edges | ( | Tree const & | tree, |
| std::vector< tree::TreeNode const * > const & | nodes | ||
| ) |
Definition at line 371 of file tree/bipartition/functions.cpp.
| utils::SvgDocument genesis::tree::get_color_tree_svg_doc_ | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::vector< utils::SvgGroup > const & | node_shapes, | ||
| std::vector< utils::SvgGroup > const & | edge_shapes | ||
| ) |
Definition at line 181 of file tree/drawing/functions.cpp.
| utils::SvgDocument get_color_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< double > const & | value_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm | ||
| ) |
Definition at line 251 of file tree/drawing/functions.cpp.
| utils::SvgDocument get_color_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch | ||
| ) |
Definition at line 238 of file tree/drawing/functions.cpp.
| utils::SvgDocument get_color_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::vector< utils::Color > const & | color_list, | ||
| std::vector< std::string > const & | color_labels | ||
| ) |
Definition at line 325 of file tree/drawing/functions.cpp.
| utils::SvgDocument get_color_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm | ||
| ) |
Definition at line 263 of file tree/drawing/functions.cpp.
| utils::SvgDocument get_color_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::vector< utils::SvgGroup > const & | node_shapes, | ||
| std::vector< utils::SvgGroup > const & | edge_shapes | ||
| ) |
Definition at line 276 of file tree/drawing/functions.cpp.
| std::vector< size_t > get_subtree_edges | ( | TreeLink const & | subtree | ) |
Definition at line 192 of file tree/bipartition/functions.cpp.
| utils::SvgDocument get_tree_svg_document | ( | CommonTree const & | tree, |
| LayoutParameters const & | params | ||
| ) |
Definition at line 226 of file tree/drawing/functions.cpp.
| utils::SvgDocument heat_tree | ( | HeatTreeParameters const & | params | ) |
Definition at line 359 of file heat_tree.cpp.
| utils::SvgDocument heat_tree | ( | HeatTreeParameters const & | params, |
| utils::ColorMap const & | matrix_color_map, | ||
| utils::ColorNormalization const & | matrix_color_norm | ||
| ) |
Definition at line 370 of file heat_tree.cpp.
| utils::SvgDocument heat_tree | ( | HeatTreeParameters const & | params, |
| utils::ColorMap const & | matrix_color_map, | ||
| utils::ColorNormalization const & | matrix_color_norm, | ||
| utils::ColorMap const & | tree_color_map, | ||
| utils::ColorNormalization const & | tree_color_norm | ||
| ) |
Definition at line 383 of file heat_tree.cpp.
| void genesis::tree::heat_tree_add_heat_matrix_bmp_ | ( | HeatTreeParameters const & | params, |
| RectangularLayout const & | layout, | ||
| utils::Matrix< utils::Color > const & | matrix, | ||
| utils::SvgDocument & | svg_doc, | ||
| HeatTreeGrid & | grid | ||
| ) |
Definition at line 283 of file heat_tree.cpp.
| void genesis::tree::heat_tree_add_heat_matrix_svg_ | ( | HeatTreeParameters const & | params, |
| RectangularLayout const & | layout, | ||
| utils::Matrix< utils::Color > const & | matrix, | ||
| utils::SvgDocument & | svg_doc, | ||
| HeatTreeGrid & | grid | ||
| ) |
Definition at line 254 of file heat_tree.cpp.
| void genesis::tree::heat_tree_add_matrix_color_scale_ | ( | utils::ColorMap const & | matrix_color_map, |
| utils::ColorNormalization const & | matrix_color_norm, | ||
| utils::SvgDocument & | svg_doc, | ||
| HeatTreeGrid & | grid | ||
| ) |
Definition at line 323 of file heat_tree.cpp.
| void genesis::tree::heat_tree_add_tree_color_scale_ | ( | RectangularLayout const & | layout, |
| utils::ColorMap const & | tree_color_map, | ||
| utils::ColorNormalization const & | tree_color_norm, | ||
| utils::SvgDocument & | svg_doc, | ||
| HeatTreeGrid & | grid | ||
| ) |
Definition at line 214 of file heat_tree.cpp.
| utils::Matrix<T> genesis::tree::heat_tree_reorder_rows_ | ( | utils::Matrix< T > const & | mat, |
| std::vector< size_t > const & | order | ||
| ) |
Definition at line 121 of file heat_tree.cpp.
|
static |
Definition at line 53 of file heat_tree.cpp.
| RectangularLayout genesis::tree::heat_tree_tree_layout_ | ( | HeatTreeParameters const & | params | ) |
Definition at line 167 of file heat_tree.cpp.
| double height | ( | Tree const & | tree | ) |
Get the height of the tree, i.e., the longest distance from the root to a leaf, measured using the branch_length.
Definition at line 169 of file tree/common_tree/functions.cpp.
| bool identical_topology | ( | std::vector< Tree > const & | trees, |
| bool | identical_indices = true |
||
| ) |
Return whether all trees have an identical topology.
See identical_topology( Tree const&, Tree const&, bool ) for details.
Definition at line 194 of file tree/function/operators.cpp.
Return whether both trees have an identical topology.
The topology is considered identical only if the order of edges is also the same in both trees. This means, although two trees might have the same number of leaves and branches, they might still be not identical (with respect to this function) when the branches appear in a different order or when the root sits at a different node.
Futhermore, if identical_indices is set to true (default), the function also checks whether the indices of nodes and edges are identical at the different nodes and edges of the trees. This is important if mulitple (identical) trees are used in a calculation, where the indices are used as indices into arrays or the like.
Definition at line 169 of file tree/function/operators.cpp.
| bool identical_topology | ( | TreeSet const & | tree_set | ) |
Return true iff all Trees in a TreeSet have an identical topology.
Definition at line 92 of file tree_set.cpp.
| size_t inner_edge_count | ( | Tree const & | tree | ) |
Return the number of Edges of a Tree that do not lead to a leaf Node.
Definition at line 201 of file tree/function/functions.cpp.
| std::vector< size_t > inner_edge_indices | ( | Tree const & | tree | ) |
Get a list of the edge indices of all inner edges, that is, all TreeEdges that do not lead to a leaf TreeNode.
Definition at line 217 of file tree/function/functions.cpp.
| size_t inner_node_count | ( | Tree const & | tree | ) |
Count the number of inner Nodes.
Definition at line 180 of file tree/function/functions.cpp.
| std::vector< size_t > inner_node_indices | ( | Tree const & | tree | ) |
Get a list of the node indices of all inner TreeNodes.
Definition at line 239 of file tree/function/functions.cpp.
| bool is_bifurcating | ( | Tree const & | tree, |
| bool | loose = false |
||
| ) |
Return whether the Tree is bifurcating.
A tree is bifurcating iff all inner nodes have exactly three neighbouring nodes. The only exception is the root node, which for rooted trees only has two neighbors. Thus, this node is allowed to have two (for rooted trees) or three (for trees with a top-level trifurcation instead of an actual root) neighbors. In order to test which of these is the case, use is_rooted().
If loose is set to true, the definition is a bit broadened and also allows other nodes with two neighbors. Such nodes do not have a furcation at all, and thus "sit" in between two other nodes. This case is however rare in practice, but can happen e.g., when wrongly rerooting the tree (in case that the old root is not deleted), or on trees that are created from a taxonomy.
Definition at line 134 of file tree/function/functions.cpp.
| bool is_binary | ( | Tree const & | tree, |
| bool | loose | ||
| ) |
Alias for is_bifurcating().
Definition at line 158 of file tree/function/functions.cpp.
| bool is_inner | ( | TreeEdge const & | edge | ) |
Return true iff the secondary node (outwards) of the given edge is an inner node.
Definition at line 84 of file tree/function/functions.cpp.
| bool is_inner | ( | TreeLink const & | link | ) |
Return true iff the node of the given link is an inner node.
Definition at line 74 of file tree/function/functions.cpp.
| bool is_inner | ( | TreeNode const & | node | ) |
Return whether the node is an inner node.
Definition at line 79 of file tree/function/functions.cpp.
| bool is_leaf | ( | TreeEdge const & | edge | ) |
Return true iff the secondary node (outwards) of the given edge is a leaf node.
Definition at line 69 of file tree/function/functions.cpp.
| bool is_leaf | ( | TreeLink const & | link | ) |
Return true iff the node of the given link is a leaf node.
Definition at line 59 of file tree/function/functions.cpp.
| bool is_leaf | ( | TreeNode const & | node | ) |
Return whether the node is a leaf/tip.
Definition at line 64 of file tree/function/functions.cpp.
| bool is_root | ( | TreeLink const & | link | ) |
Return whether the link belongs to the root node of its Tree.
Definition at line 89 of file tree/function/functions.cpp.
| bool is_root | ( | TreeNode const & | node | ) |
Return whether the node is the root of its Tree.
Definition at line 94 of file tree/function/functions.cpp.
| bool is_rooted | ( | Tree const & | tree | ) |
Return whether the Tree is rooted, that is, whether the root node has two neighbors.
Definition at line 163 of file tree/function/functions.cpp.
| void ladderize | ( | Tree & | tree, |
| LadderizeOrder | order = LadderizeOrder::kSmallFirst |
||
| ) |
Ladderize a Tree, that is, order its subtrees by size.
The function flips the TreeLink order of all internal TreeNodes of the Tree so that always the smalles/largest subtree (in number of nodes) comes first when iterating the Tree. This assumes a rooting, as the direction of the subtree of a node is measured away from the root.
Definition at line 840 of file tree/function/manipulation.cpp.
| std::vector<std::pair< TreeNode const*, double > > genesis::tree::leaf_distance_vector | ( | Tree const & | tree, |
| utils::Matrix< double > const & | node_distances, | ||
| Comparator | comp | ||
| ) |
Local helper function to calculate either closest_leaf_distance_vector() or furthest_leaf_distance_vector().
Definition at line 300 of file tree/common_tree/distances.cpp.
| size_t leaf_edge_count | ( | Tree const & | tree | ) |
Return the number of Edges of a Tree that lead to a leaf Node.
Definition at line 190 of file tree/function/functions.cpp.
| std::vector< size_t > leaf_edge_indices | ( | Tree const & | tree | ) |
Get a list of the edge indices of all leaf edges, that is, all TreeEdges that lead to a leaf TreeNode.
Definition at line 228 of file tree/function/functions.cpp.
| utils::Bitvector leaf_node_bitvector | ( | Tree const & | tree, |
| std::vector< TreeNode const * > | leaf_nodes | ||
| ) |
Return a Bitvector that has as many entries as the tree has leaf nodes, and is true where the given leaf_nodes are.
Definition at line 176 of file tree/bipartition/functions.cpp.
| size_t leaf_node_count | ( | Tree const & | tree | ) |
Count the number of leaf Nodes of a Tree.
Definition at line 168 of file tree/function/functions.cpp.
| std::vector< size_t > leaf_node_indices | ( | Tree const & | tree | ) |
Get a list of the node indices of all leaf TreeNodes.
Definition at line 250 of file tree/function/functions.cpp.
| double length | ( | Tree const & | tree | ) |
Get the length of the tree, i.e., the sum of all branch lengths.
Definition at line 160 of file tree/common_tree/functions.cpp.
| utils::Range< IteratorLevelorder< false > > genesis::tree::levelorder | ( | ElementType & | element | ) |
Definition at line 270 of file tree/iterator/levelorder.hpp.
| utils::Range< IteratorLevelorder< true > > genesis::tree::levelorder | ( | ElementType const & | element | ) |
Definition at line 260 of file tree/iterator/levelorder.hpp.
Return the lowest common ancestor of two TreeNodes.
Definition at line 649 of file tree/function/functions.cpp.
Return the lowest common ancestor of two TreeNodes.
Definition at line 613 of file tree/function/functions.cpp.
| utils::Matrix< size_t > lowest_common_ancestors | ( | Tree const & | tree | ) |
Return the lowest common ancestor of each pair of TreeNodes for a given tree, in form of a Matrix of their indices.
The entries in the resulting Matrix are the Node indices of the lowest common ancestor (LCA) of a given pair of Nodes. For example, for the result Matrix r, the entry r[ 3, 5 ] == 7 means that the LCA of Nodes 3 and 5 is Node 7.
These Nodes can for example be accesses via Tree::node_at().
Definition at line 656 of file tree/function/functions.cpp.
| TreeNode & make_rooted | ( | Tree & | tree, |
| std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> | adjust_edges = {} |
||
| ) |
Root a Tree on the first TreeEdge of its current top level TreeNode.
The tree is expected to be unrooted. The function then roots the tree on the edge that is at the primary link of the current top level trifurcation. That is, a new node is inserted on that edge, see make_rooted( Tree&, TreeEdge&, ... ) for details.
Definition at line 761 of file tree/function/manipulation.cpp.
| TreeNode & make_rooted | ( | Tree & | tree, |
| TreeEdge & | target_edge, | ||
| std::function< void(TreeEdge &target_edge, TreeEdge &new_edge)> | adjust_edges = {} |
||
| ) |
Root a Tree at a given TreeEdge.
The function expects an unrooted tree with a top-level tri- or multifurcation, which is checked via is_rooted(). It then adds a new Node that splits the given target_edge in two, and roots the tree on that new Node.
The function simply combines add_new_node( Tree&, TreeEdge& ) and change_rooting( Tree&, TreeNode& ). See there for details, and for the usage of adjust_edges.
Definition at line 747 of file tree/function/manipulation.cpp.
| Tree make_tree_from_parents_table | ( | std::vector< std::string > const & | child_names, |
| std::vector< std::string > const & | parent_names | ||
| ) |
Create a tree, given lists of child parent pairs.
The two provided lists need to have the same length, and corresponding entries that form child-parent-pairs of node names. These names are then used to create a tree.
Definition at line 170 of file tree/formats/table/reader.cpp.
| void genesis::tree::make_tree_from_parents_table_add_edge_ | ( | TreeTableHelpers & | helpers, |
| std::string const & | child_name, | ||
| std::string const & | parent_name, | ||
| Tree & | tree | ||
| ) |
Definition at line 60 of file tree/formats/table/reader.cpp.
| void make_unrooted | ( | Tree & | tree, |
| std::function< void(TreeEdge &remaining_edge, TreeEdge &deleted_edge)> | adjust_edges = {} |
||
| ) |
Unroot a Tree.
The tree is expected to be rooted, which is checked via is_rooted(). The function then removes this root by calling delete_linear_node(). See there for details, and for the usage of adjust_edges.
Definition at line 768 of file tree/function/manipulation.cpp.
| std::vector< double > mass_balance | ( | BalanceData const & | data, |
| std::unordered_set< size_t > const & | numerator_edge_indices, | ||
| std::unordered_set< size_t > const & | denominator_edge_indices | ||
| ) |
Calculate the balance of edge masses between two sets of edges.
The function calculates all balances (for all trees) between the two sets of edges.
Definition at line 357 of file balances.cpp.
| double mass_balance | ( | BalanceData const & | data, |
| std::unordered_set< size_t > const & | numerator_edge_indices, | ||
| std::unordered_set< size_t > const & | denominator_edge_indices, | ||
| size_t | tree_index | ||
| ) |
Calculate the balance of edge masses between two sets of edges.
The function calculates the balances of a specific tree between the two sets of edges. The given tree_index corresponds to the index in the original vector of MassTrees used in mass_balance_data(). This index is thus also the row index in the BalanceData::edge_masses matrix.
Definition at line 372 of file balances.cpp.
| BalanceData mass_balance_data | ( | MassTree const & | tree, |
| BalanceSettings | settings = {} |
||
| ) |
Calculate the data needed to calculate balances of a MassTree.
This is the version for a single MassTree. Here, the taxon weights are left empty, as they cannot be estimated from just a single tree. See mass_balance_data( std::vector<MassTree> const&, ... ) for the version for multiple MassTrees, which also calculates proper taxon weights.
Definition at line 337 of file balances.cpp.
| BalanceData mass_balance_data | ( | std::vector< MassTree > const & | trees, |
| BalanceSettings | settings = {} |
||
| ) |
Calculate the data needed to calculate balances on MassTrees.
This function takes a set of MassTrees and BalanceSettings and calculates the data needed for calculations such as mass_balance(), phylogenetic_ilr_transform(), and phylogenetic_factorization().
The function assumes that the Trees are populated with masses along thir branches which represent metagenomic sequences being placed there. The masses must not be normalized - there has to be at least a mass of ~1.0 per Tree for this function to work. That is each sequence should contribute about 1.0 mass to the tree (leaving out some lower masses in case of phylogenetic placement data).
Definition at line 64 of file balances.cpp.
| double mass_tree_binify_masses | ( | MassTree & | tree, |
| size_t | number_of_bins | ||
| ) |
Accumulate all masses of a MassTree into bins on each branch.
Each branch is divided into intervals of equal size, where number_of_bins is the number of those intervals. The mid points of these intervals are then used as bins, to which the masses on the branch are moved. Each mass point is moved to its closest bin, so that all mass is accumulated at the bins.
This function is useful to reduce the data load of big MassTrees, without affecting the accuracy of downstream analyses too much. Using the interval mid points as bins means that masses are moved as little as possible.
Example: Given number_of_bins == 6, for a branch of length 3.6, the bins look like this:
Intervals 0.0 0.6 1.2 1.8 2.4 3.0 3.6
| | | | | | |
^ ^ ^ ^ ^ ^
Bins 0.3 0.9 1.5 2.1 2.7 3.3
The function returns the work (mass times distance) that was needed to move the masses to the bins.
Definition at line 222 of file tree/mass_tree/functions.cpp.
| double mass_tree_center_masses_on_branches | ( | MassTree & | tree | ) |
Accumulate all masses of a MassTree on the centers of their edges.
This function can be used to minimize the data load of a MassTree. It is equal to mass_tree_binify_masses() when using number_of_bins == 1.
Return the work (mass times distance) that was needed to move the masses to the centers.
Definition at line 158 of file tree/mass_tree/functions.cpp.
| double mass_tree_center_masses_on_branches_averaged | ( | MassTree & | tree | ) |
Accumulate all masses of a MassTree at the average mass position per edge.
This function is similar to mass_tree_center_masses_on_branches(), but instead of accumulating the masses at the branch center, they are accumulated at their average position on the branch.
Return the work (mass times distance) that was needed to move the masses to the centers.
Definition at line 181 of file tree/mass_tree/functions.cpp.
| void mass_tree_clear_masses | ( | MassTree & | tree | ) |
Clear all masses of a MassTree, while keeping its topology.
Definition at line 96 of file tree/mass_tree/functions.cpp.
| std::vector< double > mass_tree_mass_per_edge | ( | MassTree const & | tree | ) |
| utils::Matrix< double > mass_tree_mass_per_edge | ( | std::vector< MassTree > const & | mass_trees | ) |
| std::vector< std::pair< double, double > > mass_tree_mass_per_edge_averaged | ( | MassTree const & | tree | ) |
Return a std::vector that contains the total Mass for each edge of the given MassTree (second value), as well as the average position of that mass on the branch (in branch lengths units, first value).
The vector is indexed using the index of the edges.
Definition at line 379 of file tree/mass_tree/functions.cpp.
| MassTree mass_tree_merge_trees | ( | MassTree const & | lhs, |
| MassTree const & | rhs, | ||
| double const | scaler_lhs = 1.0, |
||
| double const | scaler_rhs = 1.0 |
||
| ) |
Merge all masses of two MassTrees into one and return it.
The two scalers can be used to weight the masses differently, if needed.
The resulting tree will have a mass of scaler_lhs * mass(lhs) + scaler_rhs * mass(rhs), which usually is not unit mass any more. Thus, if needed, call mass_tree_normalize_masses() to rescale the masses back to unit mass.
Definition at line 60 of file tree/mass_tree/functions.cpp.
| void mass_tree_merge_trees_inplace | ( | MassTree & | lhs, |
| MassTree const & | rhs, | ||
| double const | scaler_lhs = 1.0, |
||
| double const | scaler_rhs = 1.0 |
||
| ) |
Merge all masses of two MassTrees by adding them to the first MassTree.
The two scalers can be used to weight the masses differently, if needed.
The resulting tree will have a mass of scaler_lhs * mass(lhs) + scaler_rhs * mass(rhs), which usually is not unit mass any more. Thus, if needed, call mass_tree_normalize_masses() to rescale the masses back to unit mass.
Definition at line 71 of file tree/mass_tree/functions.cpp.
| void mass_tree_normalize_masses | ( | MassTree & | tree | ) |
Scale all masses of a MassTree so that they sum up to 1.0.
Definition at line 126 of file tree/mass_tree/functions.cpp.
| void mass_tree_reverse_signs | ( | MassTree & | tree | ) |
Reverse the sign of each mass point on a MassTree.
Definition at line 104 of file tree/mass_tree/functions.cpp.
| void mass_tree_scale_masses | ( | MassTree & | tree, |
| double | factor | ||
| ) |
Scale all masses of a MassTree with the multiplicative factor factor.
Definition at line 115 of file tree/mass_tree/functions.cpp.
| double mass_tree_sum_of_masses | ( | MassTree const & | tree | ) |
Return the total sum of all masses on the MassTree.
In order for the earth_movers_distance() algorithm to work properly (and give meaningful results), the total mass on the MassTrees should ideally be the same. This function can be used to check this.
Because of numerical issues however, be aware that the result might be slighly off. This is okay, as it usually is in the last digits of the double.
Definition at line 412 of file tree/mass_tree/functions.cpp.
| void mass_tree_transform_to_unit_branch_lengths | ( | MassTree & | tree | ) |
Set all branch lengths of a MassTree to 1.0, while keeping the relative position of all masses on the branches.
Definition at line 142 of file tree/mass_tree/functions.cpp.
| bool mass_tree_validate | ( | MassTree const & | tree, |
| double | valid_total_mass_difference = -1.0 |
||
| ) |
Validate the data on a MassTree.
This function returns true iff the data on the Tree is valid:
[ 0.0, branch_length ] on their respective branches.valid_total_mass_difference is not negative, the sum of all masses is also checked. It has to be close to 0.0, using the argument as the absolute allowed difference. This is useful to check whether the masses for calculating the one-argument version of the earth_movers_distance( MassTree const& ) are correct.The function stops at the first encountered invalid condition and outputs a description message of the invalid value to LOG_INFO.
| tree | MassTree to be validated. |
| valid_total_mass_difference | If set to a non-negative value, it is used as the absolute allowed difference from zero for the total sum of all masses on the tree. |
Definition at line 423 of file tree/mass_tree/functions.cpp.
| void mass_trees_make_average_branch_lengths | ( | std::vector< MassTree > & | mass_trees | ) |
Change the branch lengths of all trees to their average, and move the masses accordingly in a proportional way.
The function only is reasonable to run if all trees have identical topology, which is however not checked explicitly. Use identical_topology() for this.
Definition at line 271 of file tree/mass_tree/functions.cpp.
| size_t max_degree | ( | Tree const & | tree | ) |
Return the highest degree of the Nodes of a Tree.
If the Tree is empty, 0 is returned.
Definition at line 125 of file tree/function/functions.cpp.
| Tree genesis::tree::minimal_tree | ( | ) |
Create a minimal Tree that can be used with manipulation functions such as add_new_node() or add_new_leaf_node() to build a custom tree, including default data types at nodees and edges.
A minimal tree for our purposes consists of two TreeNodes, a TreeEdge between them, and the respective TreeLinks.
The resulting tree has default-constructed node and edge data types of the given template arugments. See minimal_tree_topology() for a version of this function that does not create any data.
Definition at line 81 of file tree/function/manipulation.hpp.
| Tree minimal_tree_topology | ( | ) |
Create a minimal Tree that can be used with manipulation functions such as add_new_node() or add_new_leaf_node() to build a custom tree.
A minimal tree for our purposes consists of two TreeNodes, a TreeEdge between them, and the respective TreeLinks.
Note that the resulting tree does not have any data types at its nodes and edges. See minimal_tree() for a version of this function that also creates these data types.
Definition at line 55 of file tree/function/manipulation.cpp.
| utils::Matrix< double > node_branch_length_distance_matrix | ( | Tree const & | tree | ) |
Return a distance matrix containing pairwise distances between all Nodes, using the branch_length of the Edges as distance measurement.
The elements of the matrix are indexed using node().index().
Definition at line 57 of file tree/common_tree/distances.cpp.
| std::vector< double > node_branch_length_distance_vector | ( | Tree const & | tree, |
| TreeNode const * | node = nullptr |
||
| ) |
Return a vector containing the distance of all nodes with respect to the given start node, where distance is measured in the sum of branch lengths between the nodes.
The vector is indexed using the node().index() for every node. Its elements give the distance of each node with respect to the given start node. The distance is the sum of branch lengths of the edges visited on the path between the two nodes.
If no Node pointer is provided, the root is taken as node.
Definition at line 121 of file tree/common_tree/distances.cpp.
| size_t node_count | ( | Tree const & | tree | ) |
Return the number of Nodes of a Tree. Same as Tree::node_count().
Definition at line 185 of file tree/function/functions.cpp.
| utils::Range< IteratorNodeLinks< false > > genesis::tree::node_links | ( | ElementType & | element | ) |
Definition at line 196 of file node_links.hpp.
| utils::Range< IteratorNodeLinks< true > > genesis::tree::node_links | ( | ElementType const & | element | ) |
Definition at line 186 of file node_links.hpp.
| std::vector< std::string > node_names | ( | std::vector< Tree > const & | tree_set, |
| bool | leaves_only = false |
||
| ) |
Returns a set of all TreeNode names of a vector of Trees or a TreeSet.
The function returns the set of all names of all Trees in the set. See node_names(...) this version of the function for details.
Definition at line 70 of file tree/common_tree/functions.cpp.
| std::vector< std::string > node_names | ( | Tree const & | tree, |
| bool | leaves_only = false |
||
| ) |
Returns a list of all TreeNode names of a Tree.
If leaves_only is set to true, nodes names of inner nodes are not included. Unnamed nodes (node.data.name == "") are always excluded. The result is not sorted, and names are as given in the Tree (including possible duplicates).
The provided Tree needs to have TreeNodes with data types deriveed from CommonNodeData.
Definition at line 52 of file tree/common_tree/functions.cpp.
| utils::Matrix< size_t > node_path_length_matrix | ( | Tree const & | tree | ) |
Return a matrix containing the pairwise depth of all nodes of the tree.
See node_path_length_vector(...) for more information.
The vector is indexed using the node().index() for every node.
Definition at line 56 of file tree/function/distances.cpp.
| std::vector< size_t > node_path_length_vector | ( | Tree const & | tree | ) |
Return a vector containing the depth of all nodes with respect to the root node.
This function calls and returns the value of node_path_length_vector(...) using the root node of the tree.
Definition at line 134 of file tree/function/distances.cpp.
Return a vector containing the depth of all nodes with respect to the given start node.
The vector is indexed using the node().index() for every node. Its elements give the depth of each node with respect to the given start node. The depth is the number of edges visited on the path between two nodes (0 for itself, 1 for immediate neighbours, etc).
Definition at line 98 of file tree/function/distances.cpp.
| utils::Matrix< signed char > node_root_direction_matrix | ( | Tree const & | tree | ) |
Calculate a Matrix that indicates the nodes on the root side of a given node.
The row and column indices of the Matrix represent TreeNode indices. Each element of the Matrix indicates whether the column node is in the subtree of the row node that contains the root (value 1), or in a subtree that does not contain the root (value -1), while the diagonale contains 0.
Definition at line 292 of file tree/function/functions.cpp.
| std::vector< size_t > node_to_leaf_map | ( | Tree const & | tree | ) |
Definition at line 110 of file tree/bipartition/functions.cpp.
| utils::Range< IteratorPath< false > > genesis::tree::path | ( | ElementType & | start, |
| ElementType & | finish | ||
| ) |
| utils::Range< IteratorPath< true > > genesis::tree::path | ( | ElementType const & | start, |
| ElementType const & | finish | ||
| ) |
| utils::Range< IteratorPathSet< false > > genesis::tree::path_set | ( | ElementType & | start, |
| ElementType & | finish, | ||
| ElementType & | lca | ||
| ) |
Definition at line 327 of file path_set.hpp.
| utils::Range< IteratorPathSet< true > > genesis::tree::path_set | ( | ElementType const & | start, |
| ElementType const & | finish, | ||
| ElementType const & | lca | ||
| ) |
Definition at line 317 of file path_set.hpp.
Helper function that finds all TreeLinks between a given TreeNode and the root of the Tree.
Both the primary_link() of the Node and the root_link() of the Tree are included in the list. The order of the list starts at the provided node and ends at the root.
Definition at line 580 of file tree/function/functions.cpp.
| std::vector< utils::Color > phylo_factor_clade_colors | ( | Tree const & | tree, |
| std::vector< PhyloFactor > const & | factors, | ||
| size_t | num_factors = 0, |
||
| PhyloFactorCladeColors | colors = {} |
||
| ) |
Return a color for each edge, indicating which factor (phylogenetic unit, clade) it belongs to.
Phylo Factorization splits the tree into multiple parts, where each factor splits away a monophyletic clade. This function colors the edges of each clade in a different color, for visualiuzation purposes.
By default, num_factors is 0, meaning that the clades of all factors are used (up to the number of max_iterations that the factorization was run with). By setting num_factors to a smaller number, only these first clades are visualized.
The visualization is done by setting the color for the secondary edges of the factor (away from the root). This is done so that nested clades (factors found within a previously split clade) are not overwritten. Thus, the first factor leaves its primary part uncolorized, for which the base_color is used.
Definition at line 112 of file phylo_factor_colors.cpp.
| std::vector< size_t > phylo_factor_edge_indices | ( | std::vector< PhyloFactor > const & | factors, |
| size_t | max_factor = std::numeric_limits< std::size_t >::max() |
||
| ) |
Get a list of all edges that have factored out by phylogenetic_factorization().
By default, all edges that are factors are returned, that is, the list of all PhyloFactor::edge_index in the input factors (in other words, the indices of the "winning" edges). If max_factor is set to a value > 0, only this many factors (the first ones) are returned. 0 is also allowed, in which case an empty vector is returned.
Definition at line 43 of file phylo_factor_colors.cpp.
| PhyloFactor phylo_factor_find_best_edge | ( | size_t | iteration, |
| BalanceData const & | data, | ||
| std::unordered_set< size_t > const & | candidate_edges, | ||
| PhyloFactorObjectiveFunction | objective | ||
| ) |
Helper function for phylogenetic_factorization() that tries all candidate edges to find the one that maximizes the objective function.
Definition at line 183 of file phylo_factor.cpp.
| std::vector< utils::Color > phylo_factor_single_factor_colors | ( | Tree const & | tree, |
| std::vector< PhyloFactor > const & | factors, | ||
| size_t | factor_index, | ||
| PhyloFactorSingleColors | colors = {} |
||
| ) |
Return a color for each edge indicating its role in a single phylogenetic factor.
The function takes one of the factors resulting from a phylogenetic_factorization(), and colorizes the edges of the tree for user output, indicating for each edge whether:
factor_index);The resulting colors can be used for visualizing a tree.
Definition at line 60 of file phylo_factor_colors.cpp.
| std::unordered_set< size_t > phylo_factor_subtree_indices | ( | Subtree const & | subtree, |
| std::unordered_set< size_t > const & | candidate_edges | ||
| ) |
Helper function for phylogenetic_factorization() to find the constrained subtrees that are split by an edge.
Helper function to get the edge indices of a particular subtree, excluding the edge that leads to it, and excluding all subtrees that are not connected to the given subtree via the candidate edges. In other words, a subtree is excluded if it is connected to the given subtree by an edge that is not in the candidate list. Consequently, the returned indices are all part of the candidates.
Definition at line 65 of file phylo_factor.cpp.
| std::vector< PhyloFactor > phylogenetic_factorization | ( | BalanceData const & | data, |
| PhyloFactorObjectiveFunction | objective, | ||
| size_t | max_iterations = 0, |
||
| std::function< void(size_t iteration, size_t max_iterations)> | log_progress = {} |
||
| ) |
Calculate the Phylogenetic Factorization (PhyloFactor) of a set of MassTrees.
This overload also provides the iteration and edge index of the current edge when computing the objective function. This is useful when some aspect of the computation in the objective function needs to be stored for later: In that case, the objective function can be provided as a lambda that uses the additional edge_index to store the results of some objective computation. The intended use case is for instance the computation of glm_fit() in the objective function, and subsequent storage of the model output, such has deviance or beta estimates, for later inspection.
Definition at line 284 of file phylo_factor.cpp.
| std::vector< PhyloFactor > phylogenetic_factorization | ( | BalanceData const & | data, |
| std::function< double(std::vector< double > const &balances)> | objective, | ||
| size_t | max_iterations = 0, |
||
| std::function< void(size_t iteration, size_t max_iterations)> | log_progress = {} |
||
| ) |
Calculate the Phylogenetic Factorization (PhyloFactor) of a set of MassTrees.
This implementation is simiar to the ideas presented in [1]. We however extend this original idea by being able to place masses on inner branches as well, instead of just the tips (OTUs).
The function expects data coming from mass_balance_data(), and an objective function that needs to be maximized for finding the next best (greedy) phylo factor. The input to this objective function are the balances for all input data points for the current edge being considered as a factor during the execution of the greedy algorithm.
Furthermore, the number of iterations can be set via max_iterations, that is, the number of phylo factors to find. By default, all possible are found, which might take too long. Currently, we do not have a stopping criterion implemented, so it is up to the user to set a reasonable value here.
Lastly, a functional for logging the progress can be set, which needs to take the current and the maximal iteration counter (0-based) and can produce some logging for this:
[]( size_t iteration, size_t max_iterations ){
LOG_DBG1 << "iteration " << (iteration + 1) << " of " << max_iterations;
}
More details on the method can be found in
[1] A. D. Washburne, J. D. Silverman, J. W. Leff, D. J. Bennett, J. L. Darcy, S. Mukherjee, N. Fierer, and L. A. David, "Phylogenetic factorization of compositional data yields lineage-level associations in microbiome datasets," PeerJ, vol. 5, p. e2969, Feb. 2017. https://doi.org/10.7717/peerj.2969
Definition at line 268 of file phylo_factor.cpp.
| utils::Matrix< double > phylogenetic_ilr_transform | ( | BalanceData const & | data, |
| bool | reverse_signs = false |
||
| ) |
Calculate the Phylogenetic Isometric Log Ratio transformation.
The balances are calculated per node of the tree, similar to [1]. We however extend this original idea by being able to place masses on inner branches as well, instead of just the tips (OTUs). The tree has to be bifurcating and rooted. The calculated balances are stored using the node indices. Their sign (order of the subtrees) is according to the TreeLink order of each TreeNode: The numerator is the first link, the denominator is the second link - unless reverse_signs is set to true, in which case this is flipped. Use sign_matrix() to get the ordering (sign) used for the subtrees.
The function expects data coming from mass_balance_data(), which can be calculated for a single tree, or for a set of trees. In the latter case, per-taxon (that is, per-edge) weights can also be calculated, see BalanceSettings for details.
[1] J. D. Silverman, A. D. Washburne, S. Mukherjee, and L. A. David, "A phylogenetic transform enhances analysis of compositional microbiota data," Elife, vol. 6, p. e21887, Feb. 2017. https://elifesciences.org/articles/21887
Definition at line 62 of file phylo_ilr.cpp.
| utils::Range< IteratorPostorder< false > > genesis::tree::postorder | ( | ElementType & | element | ) |
Definition at line 331 of file tree/iterator/postorder.hpp.
| utils::Range< IteratorPostorder< true > > genesis::tree::postorder | ( | ElementType const & | element | ) |
Definition at line 321 of file tree/iterator/postorder.hpp.
| utils::Range< IteratorPreorder< false > > genesis::tree::preorder | ( | ElementType & | element | ) |
Definition at line 274 of file tree/iterator/preorder.hpp.
| utils::Range< IteratorPreorder< true > > genesis::tree::preorder | ( | ElementType const & | element | ) |
Definition at line 264 of file tree/iterator/preorder.hpp.
| std::string print_gist | ( | Tree const & | tree, |
| long | items | ||
| ) |
Definition at line 321 of file tree/function/operators.cpp.
| std::string print_info | ( | Tree const & | tree | ) |
Definition at line 288 of file tree/function/operators.cpp.
| std::string print_info | ( | TreeEdge const & | edge | ) |
Definition at line 297 of file tree/function/operators.cpp.
| std::string print_info | ( | TreeLink const & | link | ) |
Definition at line 305 of file tree/function/operators.cpp.
| std::string print_info | ( | TreeNode const & | node | ) |
Definition at line 313 of file tree/function/operators.cpp.
| utils::Matrix< size_t > rf_distance_absolute | ( | TreeSet const & | trees | ) |
Compute the relative RF (Robinson-Foulds) distance metric between two Trees.
The function computes the unweighted relative RF distance.
This internally simply uses rf_distance_absolute(), and divides the result properly; hence, if both variants are needed (absolute and relative), it might be faster to duplicate that normalization code (simply copy from this function), instead of computing the RF distance twice, which is what happens if both rf_distance_relative() and rf_distance_absolute() are called.
Compute the relative RF (Robinson-Foulds) distance metric between a given lhs Tree and all of the trees in the rhs TreeSet.
The function computes the unweighted relative RF distance. This is meant as an accelaration if the pairwise distance is not needed.
This internally simply uses rf_distance_absolute(), and divides the result properly; hence, if both variants are needed (absolute and relative), it might be faster to duplicate that normalization code (simply copy from this function), instead of computing the RF distance twice, which is what happens if both rf_distance_relative() and rf_distance_absolute() are called.
| utils::Matrix< double > rf_distance_relative | ( | TreeSet const & | trees | ) |
Compute the pairwise relative RF (Robinson-Foulds) distance metric between a set of trees.
The function computes the unweighted relative RF distance.
This internally simply uses rf_distance_absolute(), and divides the result properly; hence, if both variants are needed (absolute and relative), it might be faster to duplicate that normalization code (simply copy from this function), instead of computing the RF distance twice, which is what happens if both rf_distance_relative() and rf_distance_absolute() are called.
| std::vector< utils::Bitvector > rf_get_bitvectors | ( | Tree const & | tree, |
| std::unordered_map< std::string, size_t > const & | names | ||
| ) |
Get all split Bitvectors for a given Tree.
For each inner edge of the tree, a Bitvector is produced that contains true bits at all indices of the tips on one side of the split, using names for getting indices of leaf nodes.
The Bitvectors are normalized, that is, their first bit is always set to 0. This makes sure that the two ways of representing each split result in the same Bitvector.
| void genesis::tree::rf_get_bitvectors_template | ( | Tree const & | tree, |
| std::unordered_map< std::string, size_t > const & | names, | ||
| BitvectorProcessor const & | process_bitvector | ||
| ) |
Local helper function template that constructs all bitvectors for the splits of a tree, but allows to customize what to do with them once constructed.
This is meant to avoid code duplication, but still gives performant code due to the local scope of the template: if the compiler is halfway smart, it will inline everything.
| std::unordered_map< utils::Bitvector, utils::Bitvector > rf_get_occurrences | ( | Tree const & | lhs, |
| Tree const & | rhs | ||
| ) |
Get an occurrence map for each split found in two trees.
This is a special case of the more general rf_get_occurrences( TreeSet const& trees ) function, which takes two Trees and computes their split occurences.
| std::unordered_map< utils::Bitvector, utils::Bitvector > rf_get_occurrences | ( | Tree const & | lhs, |
| TreeSet const & | rhs | ||
| ) |
Get an occurrence map for each split found in some Trees.
This is a special case of the more general rf_get_occurrences( TreeSet const& trees ) function, which takes one additional Tree into account. This lhs tree gets index 0 in the resulting Bitvectors of the mapped values, while all trees in rhs get their index in the TreeSet plus one.
The function is meant as an accelaration for computing the distance from one Tree to several other Trees, and is used by rf_distance_absolute( Tree const& lhs, TreeSet const& rhs ).
| std::unordered_map< utils::Bitvector, utils::Bitvector > rf_get_occurrences | ( | TreeSet const & | trees | ) |
Get an occurrence map for each split found in the given TreeSet.
The given trees need to contain the same leaf node names. Then, all their splits are computed, represented a Bitvectors of tips. For each such split, another Bitvector is produced that is true at every tree in trees that contains that split.
In other words, the function yields a map from Bitvectors (keys) that represent splits to Bitvectors (mapped values) that represent occurences of these splits in the given trees. The size of the map hence is the number of unique splits in all trees; the size of the key Bitvectors is the number of taxa in the trees; and the size of the mapped value Bitvectors is the number of trees, that is TreeSet::size().
| std::unordered_map< std::string, size_t > rf_taxon_name_map | ( | Tree const & | tree | ) |
Get a mapping from taxon names to unique IDs.
The IDs are consecutive, starting at 0. Only tip/leaf names are processed, which need to be unique and non-empty.
The mappning can then be used for unique identification of taxa, which is needed for the RF distance calculation functions.
| void scale_all_branch_lengths | ( | Tree & | tree, |
| double | factor = 1.0 |
||
| ) |
Scale all branch lengths of a Tree by a given factor.
This function simply multiplies all branch lengths with the given factor. See also set_all_branch_lengths() for setting the branch lengths to a value.
Definition at line 233 of file tree/common_tree/functions.cpp.
| void set_all_branch_lengths | ( | Tree & | tree, |
| double | length = 1.0 |
||
| ) |
Set all branch lengths of a Tree to a given value.
See also scale_all_branch_lengths() for a scaling function.
Definition at line 224 of file tree/common_tree/functions.cpp.
| utils::Matrix< signed char > sign_matrix | ( | Tree const & | tree, |
| bool | compressed = false |
||
| ) |
Compute the sign matrix or Sequential Binary Partition (SBP) of a Tree.
The Tree has to be rooted and strictly bifurcating. Then, we can compute a matrix that tells for each node a relative ordering of the other nodes. Say we have the tree:
/----------- T3 /-----| N2 | \----------- T2 | N1 | \----------- T1
This yields the (compressed) sign matrix:
| Taxa | T1 | T2 | T3 |
|---|---|---|---|
| N1 | +1 | -1 | -1 |
| N2 | 0 | +1 | -1 |
That is, all nodes in the subtree of the first child of a node get assigned a +1, and all nodes in the subtree of the second child get a -1. The remaining nodes (the rest of the tree, including the node itself) get assigned a 0.
This was introduced as Sequential Binary Partition (SBP) in [1], and called Sign Matrix in [2]. See there for more details.
By default, the matrix dimensions are n*n with n being the number of nodes in the tree. That is, all node indices are used. This is easiest to work with in the context of other functions that used node indices. However, the matrix contains a lot of zeros.
If however compressed is true, the matrix is not indexed by the node indices. Instead, the rows only contain inner nodes, and the columns only contain leaf nodes, as shown in the table above. That means that the sign of inner nodes is not available in the matrix. Use inner_node_indices() to get the node indices that correspond to the rows of the matrix, and use leaf_node_indices() for the node indices that correspond to the columns.
[1] V. Pawlowsky-Glahn and J. J. Egozcue, "Exploring Compositional Data with the CoDa-Dendrogram," Austrian J. Stat., vol. 40, no. 1&2, pp. 103–113, 2011. https://ajs.or.at/index.php/ajs/article/view/vol40,%20no1&2%20-%2011
[2] J. D. Silverman, A. D. Washburne, S. Mukherjee, and L. A. David, "A phylogenetic transform enhances analysis of compositional microbiota data," Elife, vol. 6, p. e21887, Feb. 2017. https://elifesciences.org/articles/21887
Definition at line 505 of file tree/function/functions.cpp.
Calculate the height of a subtree, that is, the maximum path length to a leaf of that subtree, measured in edges between the link and the leaf.
Definition at line 440 of file tree/function/functions.cpp.
| std::vector< size_t > subtree_max_path_heights | ( | Tree const & | tree | ) |
Definition at line 500 of file tree/function/functions.cpp.
Definition at line 461 of file tree/function/functions.cpp.
Return the size of the subtree defined by the given TreeLink, measured in number of nodes.
Definition at line 345 of file tree/function/functions.cpp.
| std::vector< size_t > subtree_sizes | ( | Tree const & | tree | ) |
Calculate the sizes of all subtrees as seen from the root of the tree.
See subtree_sizes(...) for details.
Definition at line 435 of file tree/function/functions.cpp.
Calculate the sizes of all subtrees as seen from the given TreeNode.
The function returns a vector with as many elements as the Tree has nodes. The vector is indexed using the TreeNode::index() values.
Each value in the vector tells the size (in number of nodes) of the subtree of the correnspinding node, as seen from the given starting node, and excluding that starting node.
In methaphorical words, the given starting node is used as a hook where the tree is suspended from, so that it hangs down. A subtree is then the part of the tree that "hangs down" from a certain node. We then count the number of nodes in each of those subtrees (that is, we examine the subtree starting at each node of the tree). For the starting node, the count is always the number of nodes of the tree minus one (because the node is not counted itself).
Definition at line 367 of file tree/function/functions.cpp.
| bool genesis::tree::tree_data_is | ( | Tree const & | tree, |
| bool | allow_null = false |
||
| ) |
Check whether the data of the nodes and edges of the Tree are exactly of the specified data types.
This function returns true iff all data types of the nodes and edges match exaclty the specified types. It uses typeid() for this matching. If allow_null is set to true, data pointers are allowed be empty (null_ptr).
Definition at line 66 of file tree/function/operators.hpp.
| bool genesis::tree::tree_data_is_derived_from | ( | Tree const & | tree, |
| bool | allow_null = false |
||
| ) |
Check whether the data of the nodes and edges of the Tree are derived from the specified data types.
This function returns true iff all data types of the nodes and edges are derived from the specified types. It uses dynamic_cast() for this. If allow_null is set to true, data pointers are allowed be empty (null_ptr).
Definition at line 112 of file tree/function/operators.hpp.
| bool validate_topology | ( | Tree const & | tree | ) |
Validate that all internal pointers of the Tree elements (TreeLinks, TreeNodes, TreeEdges) to each other are correct and that some other invariants are met.
Validate the correctness of all Tree pointers etc.
This check is a bit pedantic, but better safe than sorry.
Definition at line 332 of file tree/function/operators.cpp.
| void write_color_tree_to_nexus_file | ( | CommonTree const & | tree, |
| std::vector< double > const & | value_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::string const & | nexus_filename | ||
| ) |
Definition at line 165 of file tree/drawing/functions.cpp.
| void write_color_tree_to_nexus_file | ( | CommonTree const & | tree, |
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::string const & | nexus_filename | ||
| ) |
Definition at line 131 of file tree/drawing/functions.cpp.
| void write_color_tree_to_phyloxml_file | ( | CommonTree const & | tree, |
| std::vector< double > const & | value_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::string const & | phyloxml_filename | ||
| ) |
Definition at line 106 of file tree/drawing/functions.cpp.
| void write_color_tree_to_phyloxml_file | ( | CommonTree const & | tree, |
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::string const & | phyloxml_filename | ||
| ) |
Definition at line 88 of file tree/drawing/functions.cpp.
| void write_color_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< double > const & | value_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 402 of file tree/drawing/functions.cpp.
| void write_color_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 386 of file tree/drawing/functions.cpp.
| void write_color_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| std::vector< utils::Color > const & | color_list, | ||
| std::vector< std::string > const & | color_labels, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 451 of file tree/drawing/functions.cpp.
| void write_color_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 415 of file tree/drawing/functions.cpp.
| void write_color_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::vector< utils::Color > const & | color_per_branch, | ||
| utils::ColorMap const & | color_map, | ||
| utils::ColorNormalization const & | color_norm, | ||
| std::vector< utils::SvgGroup > const & | node_shapes, | ||
| std::vector< utils::SvgGroup > const & | edge_shapes, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 432 of file tree/drawing/functions.cpp.
| void write_tree_to_newick_file | ( | CommonTree const & | tree, |
| std::string const & | newick_filename | ||
| ) |
Write a newick file containing a tree.
This is a very simple wrapper for common cases.
Definition at line 68 of file tree/drawing/functions.cpp.
| void write_tree_to_nexus_file | ( | CommonTree const & | tree, |
| std::string const & | nexus_filename | ||
| ) |
Write a nexus file containing a tree.
The file format can be read and visualized by, e.g., FigTree.
Definition at line 122 of file tree/drawing/functions.cpp.
| void write_tree_to_phyloxml_file | ( | CommonTree const & | tree, |
| std::string const & | phyloxml_filename | ||
| ) |
Write a phyloxml file containing a tree.
The file format can be read and visualized by, e.g., Archaeopteryx.
Definition at line 79 of file tree/drawing/functions.cpp.
| void write_tree_to_svg_file | ( | CommonTree const & | tree, |
| LayoutParameters const & | params, | ||
| std::string const & | svg_filename | ||
| ) |
Definition at line 371 of file tree/drawing/functions.cpp.
| using AttributeTree = Tree |
Alias for a Tree that stores TreeNodes and TreeEdges with string attributes on them.
Definition at line 51 of file tree/attribute_tree/tree.hpp.
| using AttributeTreeEdge = TreeEdge |
Alias for a TreeEdge of an AttributeTree. See there for more information.
Definition at line 57 of file tree/attribute_tree/tree.hpp.
| using AttributeTreeLink = TreeLink |
Alias for a TreeLink of an AttributeTree. See there for more information.
Definition at line 63 of file tree/attribute_tree/tree.hpp.
| using AttributeTreeMap = std::map<std::string, std::string> |
Alias for the map type used by an AttributeTree.
We define this alias in the namespace instead of inside of a class, because it is used in multiple places. Defining it here once allows to easily change the type in the future, should that be needed.
See AttributeTreeNodeData and AttributeTreeEdgeData for the data classes where this type is used.
Definition at line 81 of file tree/attribute_tree/tree.hpp.
| using AttributeTreeNode = TreeNode |
Alias for a TreeNode of an AttributeTree. See there for more information.
Definition at line 69 of file tree/attribute_tree/tree.hpp.
| typedef Tree CommonTree |
Alias for a Tree with data types CommonNodeData and CommonEdgeData.
Definition at line 55 of file placement/function/distances.hpp.
| using CommonTreeEdge = TreeEdge |
Alias for a TreeEdge of a CommonTree. See there for more information.
Definition at line 53 of file tree/common_tree/tree.hpp.
| using CommonTreeLink = TreeLink |
Alias for a TreeLink of a CommonTree. See there for more information.
Definition at line 58 of file tree/common_tree/tree.hpp.
| using CommonTreeNode = TreeNode |
Alias for a TreeNode of a CommonTree. See there for more information.
Definition at line 63 of file tree/common_tree/tree.hpp.
| using LayoutTree = tree::Tree |
Alias for a tree::Tree used for a tree with information needed for tree drawing.
Definition at line 51 of file layout_tree.hpp.
| using LayoutTreeEdge = tree::TreeEdge |
Alias for tree::TreeEdge used in a LayoutTree. See LayoutEdgeData for the data stored on the edges.
Definition at line 63 of file layout_tree.hpp.
| using LayoutTreeLink = tree::TreeLink |
Alias for tree::TreeLink used in a LayoutTree.
Definition at line 68 of file layout_tree.hpp.
| using LayoutTreeNode = tree::TreeNode |
Alias for tree::TreeNode used in a LayoutTree. See LayoutNodeData for the data stored on the nodes.
Definition at line 57 of file layout_tree.hpp.
Alias for a Tree that stores masses on its TreeEdges.
It is for example used to calculate the earth movers distance between to sets of masses distributed on a Tree.
See earth_movers_distance( MassTree const& tree, double ) for more details on the purpose of this tree type and on the earth movers distance in general.
The branches of an MassTree hold a list of masses, sorted along their position on the branch.
It is easily possible to merge the masses of two MassTrees by using mass_tree_merge_trees() or mass_tree_merge_trees_inplace().
Lastly, there are some some useful transformation functions:
See there for details.
Definition at line 54 of file placement/function/operators.hpp.
| using MassTreeEdge = TreeEdge |
Alias for a TreeEdge of a MassTree. See there for more information.
Definition at line 76 of file tree/mass_tree/tree.hpp.
| using MassTreeLink = TreeLink |
Alias for a TreeLink of a MassTree. See there for more information.
Definition at line 81 of file tree/mass_tree/tree.hpp.
| using MassTreeNode = TreeNode |
Alias for a TreeNode of a MassTree. See there for more information.
Definition at line 86 of file tree/mass_tree/tree.hpp.
| using PhyloFactorObjectiveFunction = std::function< double( size_t iteration, size_t edge_index, std::vector<double> const& balances ) > |
Function type used as the objective function in phylogenetic_factorization().
Definition at line 61 of file phylo_factor.hpp.
| using SimpleNewickTree = Tree |
Alias for a Tree that stores TreeNodes and TreeEdges with the standard Newick elements on them.
This tree contains node and edge data which are derived from the CommonTreeNodeData and CommonTreeEdgeData, respectively, but additionally contains support for all other elements that can occur in a Newick file format tree:
:[bootstrap]:[prob] fields after the branch length.[], which are also often (mis-)used for ad-hoc and more established extensions such as the New Hampshire eXtended (NHX) format [&&NHX:key=value:...].{1}.With additional data are stored in the tree nodes (comments) and edges (branch values and jplace tags), respectively.
Hence, this tree type is also the most direct representation of our internal NewickBrokerElement data, which we use for parsing Newick files in the NewickReader.
Definition at line 70 of file simple_tree.hpp.
| using SimpleNewickTreeEdge = TreeEdge |
Alias for a TreeEdge of an SimpleNewickTree. See there for more information.
Definition at line 76 of file simple_tree.hpp.
| using SimpleNewickTreeLink = TreeLink |
Alias for a TreeLink of an SimpleNewickTree. See there for more information.
Definition at line 82 of file simple_tree.hpp.
| using SimpleNewickTreeNode = TreeNode |
Alias for a TreeNode of an SimpleNewickTree. See there for more information.
Definition at line 88 of file simple_tree.hpp.
|
strong |
| Enumerator | |
|---|---|
| kSmallFirst | |
| kLargeFirst | |
Definition at line 413 of file tree/function/manipulation.hpp.
|
strong |
Shape of the tree for drawing, either circular or rectangular.
| Enumerator | |
|---|---|
| kCircular | |
| kRectangular | |
Definition at line 50 of file layout_base.hpp.
|
strong |
Spreading of the nodes of a tree for drawing.
In tree drawing, one axis is usually used for the branch lengths (or at least, for distancing nodes from each other in a cladegram), while the other axis does not have a biological meaning. It is instead used to spread out the nodes so that the tree is actually drawn in a plane instead of just a line.
Using this setting, the spreading can be controlled: Default is to spread out the leaves evenly, giving the typical tree layout. Sometimes however it is necessary to also make space for inner nodes. This is what the other options are for (with or without the root as a special case).
| Enumerator | |
|---|---|
| kLeafNodesOnly | |
| kAllNodesButRoot | |
| kAllNodes | |
Definition at line 80 of file layout_base.hpp.
|
strong |
Type of tree for drawing, either phylogram or cladogram.
A phylogram uses and shows branch lengths, while a cladegram aligns all leaf nodes to each other, and adjusts inner nodes accordingly.
| Enumerator | |
|---|---|
| kPhylogram | |
| kCladogram | |
Definition at line 62 of file layout_base.hpp.