Classes | |
| struct | EpcaData |
| Helper stucture that collects the output of epca(). More... | |
| class | JplaceReader |
| Read Jplace data. More... | |
| class | JplaceWriter |
| Write Jplace data. More... | |
| struct | NodeDistanceHistogram |
| Simple histogram data structure with equal sized bins. More... | |
| struct | NodeDistanceHistogramSet |
| Collection of NodeDistanceHistograms that describes one Sample. More... | |
| class | PlacementEdgeData |
Data class for PlacementTreeEdges. Stores the branch length of the edge, and the edge_num, as defined in the jplace standard. More... | |
| class | PlacementNodeData |
| Data class for PlacementTreeNodes. Stores a node name. More... | |
| class | PlacementTreeNewickReader |
| class | PlacementTreeNewickReaderPlugin |
| class | PlacementTreeNewickWriter |
| class | PlacementTreeNewickWriterPlugin |
| class | Pquery |
| A pquery holds a set of PqueryPlacements and a set of PqueryNames. More... | |
| class | PqueryName |
| A name of a Pquery and its multiplicity. More... | |
| class | PqueryPlacement |
| One placement position of a Pquery on a Tree. More... | |
| struct | PqueryPlacementPlain |
| Simple POD struct for a Placement used for speeding up some calculations. More... | |
| struct | PqueryPlain |
| Simple POD struct that stores the information of a Pquery in a simple format for speeding up some calculations. More... | |
| class | Sample |
| Manage a set of Pqueries along with the PlacementTree where the PqueryPlacements are placed on. More... | |
| class | SampleSerializer |
| class | SampleSet |
| Store a set of Samples with associated names. More... | |
| class | Simulator |
| Simulate Pqueries on the Tree of a Sample. More... | |
| class | SimulatorEdgeDistribution |
| class | SimulatorExtraPlacementDistribution |
| Generate a certain number of additional PqueryPlacements around a given PlacementTreeEdge. More... | |
| class | SimulatorLikeWeightRatioDistribution |
| class | SimulatorPendantLengthDistribution |
| class | SimulatorProximalLengthDistribution |
Functions | |
| double | add_sample_to_mass_tree (Sample const &smp, double const sign, double const scaler, tree::MassTree &target) |
| Helper function to copy masses from a Sample to a MassTree. More... | |
| void | adjust_branch_lengths (Sample &sample, tree::Tree const &source) |
Take the branch lengths of the source Tree and use them as the new branch lengths of the sample. More... | |
| void | adjust_branch_lengths (SampleSet &sample_set, tree::Tree const &source) |
Take the branch lengths of the source Tree and use them as the new branch lengths of the Samples in the sample_set. More... | |
| void | adjust_to_average_branch_lengths (SampleSet &sample_set) |
Set the branch lengths of all Samples in the sample_set to the respecitve average branch length of the Samples. More... | |
| bool | all_identical_trees (SampleSet const &sample_set) |
| Returns true iff all Trees of the Samples in the set are identical. More... | |
| std::unordered_set< std::string > | all_pquery_names (Sample const &sample) |
| Return a set of all unique PqueryNames of the Pqueries of the given sample. More... | |
| tree::Tree | average_branch_length_tree (SampleSet const &sample_set) |
| Return the Tree that has edges with the average branch length of the respective edges of the Trees in the Samples of the given SampleSet. More... | |
| std::pair< PlacementTreeEdge const *, double > | center_of_gravity (Sample const &smp, bool const with_pendant_length=false) |
| Calculate the Center of Gravity of the placements on a tree. More... | |
| double | center_of_gravity_distance (Sample const &smp_a, Sample const &smp_b, bool const with_pendant_length=false) |
| Calculate the distance between the two Centers of Gravity of two Samples. More... | |
| double | center_of_gravity_variance (Sample const &smp, bool const with_pendant_length=false) |
| Calculate the variance of the PqueryPlacements of a Sample around its Center of Gravity. More... | |
| std::vector< int > | closest_leaf_depth_histogram (Sample const &smp) |
| Return a histogram representing how many placements have which depth with respect to their closest leaf node. More... | |
| std::vector< int > | closest_leaf_distance_histogram (Sample const &smp, const double min, const double max, const int bins=10) |
| Returns a histogram counting the number of placements that have a certain distance to their closest leaf node, divided into equally large intervals between a min and a max distance. More... | |
| std::vector< int > | closest_leaf_distance_histogram_auto (Sample const &smp, double &min, double &max, const int bins=10) |
| Returns the same type of histogram as closest_leaf_distance_histogram(), but automatically determines the needed boundaries. More... | |
| std::vector< double > | closest_leaf_weight_distribution (Sample const &sample) |
| void | collect_duplicate_pqueries (Sample &smp) |
| Find all Pqueries that share a common name and combine them into a single Pquery containing all their collective PqueryPlacements and PqueryNames. More... | |
| bool | compatible_trees (PlacementTree const &lhs, PlacementTree const &rhs) |
| Return whether two PlacementTrees are compatible. More... | |
| bool | compatible_trees (Sample const &lhs, Sample const &rhs) |
| Return whether the PlacementTrees of two Samples are compatible. More... | |
| PlacementTree | convert_common_tree_to_placement_tree (tree::CommonTree const &source_tree) |
| Convert a CommonTree into a PlacementTree. More... | |
| std::pair< tree::TreeSet, std::vector< double > > | convert_sample_set_to_mass_trees (SampleSet const &sample_set, bool normalize) |
| Convert all Samples in a SampleSet to tree::MassTrees. More... | |
| std::pair< tree::MassTree, double > | convert_sample_to_mass_tree (Sample const &sample, bool normalize) |
| Convert a Sample to a tree::MassTree. More... | |
| void | copy_pqueries (Sample const &source, Sample &target) |
| Copy all Pqueries from the source Sample (left parameter) to the target Sample (right parameter). More... | |
| double | earth_movers_distance (Sample const &lhs, Sample const &rhs, double const p=1.0, bool const with_pendant_length=false) |
| Calculate the earth mover's distance between two Samples. More... | |
| utils::Matrix< double > | earth_movers_distance (SampleSet const &sample_set, double const p=1.0, bool const with_pendant_length=false) |
| Calculate the pairwise Earth Movers Distance for all Samples in a SampleSet. More... | |
| std::unordered_map< int, PlacementTreeEdge * > | edge_num_to_edge_map (PlacementTree const &tree) |
Return a mapping of edge_num integers to the corresponding PlacementTreeEdge object. More... | |
| std::unordered_map< int, PlacementTreeEdge * > | edge_num_to_edge_map (Sample const &smp) |
| Return a mapping of edge_num integers to the corresponding PlacementTreeEdge object. More... | |
| double | edpl (Pquery const &pquery, utils::Matrix< double > const &node_distances) |
| Calculate the EDPL uncertainty values for a Pquery. More... | |
| std::vector< double > | edpl (Sample const &sample) |
| Calculate the expected distance between placement locations (EDPL) for all Pqueries in a Sample. More... | |
| double | edpl (Sample const &sample, Pquery const &pquery) |
| Calculate the EDPL uncertainty values for a Pquery. More... | |
| std::vector< double > | edpl (Sample const &sample, utils::Matrix< double > const &node_distances) |
| Calculate the edpl() for all Pqueries in the Sample. More... | |
| EpcaData | epca (SampleSet const &samples, double kappa=1.0, double epsilon=1e-5, size_t components=0) |
| Perform EdgePCA on a SampleSet. More... | |
| utils::Matrix< double > | epca_imbalance_matrix (SampleSet const &samples, bool include_leaves=false, bool normalize=true) |
| Calculate the imbalance matrix of placment mass for all Samples in a SampleSet. More... | |
| std::vector< double > | epca_imbalance_vector (Sample const &sample, bool normalize=true) |
| Calculate the imbalance of placement mass for each Edge of the given Sample. More... | |
| void | epca_splitify_transform (utils::Matrix< double > &imbalance_matrix, double kappa=1.0) |
| Perform a component-wise transformation of the imbalance matrix used for epca(). More... | |
| static void | fill_node_distance_histogram_set_ (Sample const &sample, utils::Matrix< double > const &node_distances, utils::Matrix< signed char > const &node_sides, NodeDistanceHistogramSet &histogram_set) |
| Local helper function to fill the placements of a Sample into Histograms. More... | |
| void | filter_max_pendant_length (Pquery &pquery, double threshold) |
Remove all PqueryPlacements that have a pendant_length above the given threshold. More... | |
| void | filter_max_pendant_length (Sample &sample, double threshold) |
Remove all PqueryPlacements that have a pendant_length above the given threshold from all Pqueries of the Sample. More... | |
| void | filter_min_accumulated_weight (Pquery &pquery, double threshold=0.99) |
Remove the PqueryPlacements with the lowest like_weight_ratio, while keeping the accumulated weight (sum of all remaining like_weight_ratios) above a given threshold. More... | |
| void | filter_min_accumulated_weight (Sample &smp, double threshold=0.99) |
Remove the PqueryPlacements with the lowest like_weight_ratio, while keeping the accumulated weight (sum of all remaining like_weight_ratios) above a given threshold. More... | |
| void | filter_min_pendant_length (Pquery &pquery, double threshold) |
Remove all PqueryPlacements that have a pendant_length below the given threshold. More... | |
| void | filter_min_pendant_length (Sample &sample, double threshold) |
Remove all PqueryPlacements that have a pendant_length below the given threshold from all Pqueries of the Sample. More... | |
| void | filter_min_weight_threshold (Pquery &pquery, double threshold=0.01) |
Remove all PqueryPlacements that have a like_weight_ratio below the given threshold. More... | |
| void | filter_min_weight_threshold (Sample &smp, double threshold=0.01) |
Remove all PqueryPlacements that have a like_weight_ratio below the given threshold from all Pqueries of the Sample. More... | |
| void | filter_n_max_weight_placements (Pquery &pquery, size_t n=1) |
Remove all PqueryPlacements but the n most likely ones from the Pquery. More... | |
| void | filter_n_max_weight_placements (Sample &smp, size_t n=1) |
Remove all PqueryPlacements but the n most likely ones from all Pqueries in the Sample. More... | |
| template<typename F > | |
| void | filter_pqueries_by_name_ (Sample &smp, F predicate, bool remove_empty_pqueries) |
| void | filter_pqueries_differing_names (Sample &sample_1, Sample &sample_2, bool remove_empty_name_pqueries=true) |
| Remove all PqueryNames from the two Samples that occur in both of them. More... | |
| void | filter_pqueries_intersecting_names (Sample &sample_1, Sample &sample_2, bool remove_empty_name_pqueries=true) |
| Remove all PqueryNames from the two Samples that are unique to each of them. More... | |
| void | filter_pqueries_keeping_names (Sample &smp, std::string const ®ex, bool remove_empty_name_pqueries=true) |
Remove all PqueryNames which do not match the given regex. More... | |
| void | filter_pqueries_keeping_names (Sample &smp, std::unordered_set< std::string > const &keep_list, bool remove_empty_name_pqueries=true) |
Remove all PqueryNames which do not occur in the given keep_list. More... | |
| void | filter_pqueries_removing_names (Sample &smp, std::string const ®ex, bool remove_empty_name_pqueries=true) |
Remove all PqueryNames which match the given regex. More... | |
| void | filter_pqueries_removing_names (Sample &smp, std::unordered_set< std::string > const &remove_list, bool remove_empty_name_pqueries=true) |
Remove all PqueryNames which occur in the given remove_list. More... | |
| Pquery * | find_pquery (Sample &smp, std::string const &name) |
| Return the first Pquery that has a particular name, or nullptr of none has. More... | |
| Pquery const * | find_pquery (Sample const &smp, std::string const &name) |
| Return the first Pquery that has a particular name, or nullptr of none has. More... | |
| Sample * | find_sample (SampleSet &sample_set, std::string const &name) |
Get the first Sample in a SampleSet that has a given name, or nullptr if not found. More... | |
| Sample const * | find_sample (SampleSet const &sample_set, std::string const &name) |
Get the first Sample in a SampleSet that has a given name, or nullptr if not found. More... | |
| bool | has_consecutive_edge_nums (PlacementTree const &tree) |
Verify that the PlacementTree has no duplicate edge_nums and that they form consecutive numbers starting from 0. More... | |
| bool | has_correct_edge_nums (PlacementTree const &tree) |
| Verify that the tree has correctly set edge nums. More... | |
| bool | has_name (Pquery const &pquery, std::string const &name) |
| Return true iff the given Pquery contains a particular name. More... | |
| bool | has_name (Sample const &smp, std::string const &name) |
| Return true iff the given Sample contains a Pquery with a particular name, i.e., a PqueryName whose name member equals the given name. More... | |
| tree::Tree | labelled_tree (Sample const &sample, bool fully_resolve=false, std::string const &name_prefix="") |
| Produce a Tree where the most probable PqueryPlacement of each Pquery in a Sample is turned into an Edge. More... | |
| tree::Tree | labelled_tree (Sample const &sample, tree::Tree const &tree, bool fully_resolve=false, std::string const &name_prefix="") |
| Produce a Tree where each PqueryPlacement of a Sample is turned into an Edge. More... | |
| void | labelled_tree_add_lonely_placement_ (tree::Tree &tree, tree::TreeEdge &edge, LabelledTreePlacementPair const &placement_pair, std::string const &name_prefix) |
| std::vector< std::vector< LabelledTreePlacementPair > > | labelled_tree_placement_pairs_per_edge_ (Sample const &sample) |
| void | labelled_tree_process_edge_fully_resolved_ (tree::Tree &tree, tree::TreeEdge &edge, std::vector< LabelledTreePlacementPair > const &placement_pairs, std::string const &name_prefix) |
| void | labelled_tree_process_edge_multifurcating_ (tree::Tree &tree, tree::TreeEdge &edge, std::vector< LabelledTreePlacementPair > const &placement_pairs, std::string const &name_prefix) |
| void | learn_like_weight_ratio_distribution (Sample const &sample, SimulatorLikeWeightRatioDistribution &lwr_distib, size_t number_of_intervals) |
| void | learn_per_edge_weights (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Sets the weights of an SimulatorEdgeDistributionso that they follow the same distribution of placement weight per edge as a given Sample. More... | |
| void | learn_placement_number_weights (Sample const &sample, SimulatorExtraPlacementDistribution &p_distib) |
| void | learn_placement_path_length_weights (Sample const &sample, SimulatorExtraPlacementDistribution &p_distib) |
| static NodeDistanceHistogramSet | make_empty_node_distance_histogram_set_ (tree::Tree const &tree, utils::Matrix< double > const &node_distances, utils::Matrix< signed char > const &node_sides, size_t const histogram_bins) |
| Local helper function to create a set of Histograms without any weights for a given Tree. More... | |
| void | make_rooted (Sample &sample, PlacementTreeEdge &target_edge) |
| Root the underlying PlacementTree of a Sample at a specified TreeEdge. More... | |
| Sample | merge_all (SampleSet const &sample_set) |
| Returns a Sample where all Samples of a SampleSet have been merged into. More... | |
| void | merge_duplicate_names (Pquery &pquery) |
Merge all PqueryNames that have the same name property into one, while adding up their multiplicity. More... | |
| void | merge_duplicate_names (Sample &smp) |
Call merge_duplicate_names() for each Pquery of the Sample. More... | |
| void | merge_duplicate_placements (Pquery &pquery) |
| Merge all PqueryPlacements of a Pquery that are on the same TreeEdge into one averaged PqueryPlacement. More... | |
| void | merge_duplicate_placements (Sample &smp) |
| Call merge_duplicate_placements( Pquery& ) for each Pquery of a Sample. More... | |
| void | merge_duplicates (Sample &smp) |
| Look for Pqueries with the same name and merge them. More... | |
| NodeDistanceHistogramSet | node_distance_histogram_set (Sample const &sample, utils::Matrix< double > const &node_distances, utils::Matrix< signed char > const &node_sides, size_t const histogram_bins) |
| Calculate the NodeDistanceHistogramSet representing a single Sample, given the necessary matrices of this Sample. More... | |
| static NodeDistanceHistogramSet | node_distance_histogram_set (Sample const &sample, size_t const histogram_bins) |
| static std::vector< NodeDistanceHistogramSet > | node_distance_histogram_set (SampleSet const &sample_set, size_t const histogram_bins) |
| Local helper function that calculates all Histograms for all Samples in a SampleSet. More... | |
| static double | node_histogram_distance (NodeDistanceHistogram const &lhs, NodeDistanceHistogram const &rhs) |
| double | node_histogram_distance (NodeDistanceHistogramSet const &lhs, NodeDistanceHistogramSet const &rhs) |
| Given the histogram sets that describe two Samples, calculate their distance. More... | |
| double | node_histogram_distance (Sample const &sample_a, Sample const &sample_b, size_t const histogram_bins=25) |
| Calculate the Node Histogram Distance of two Samples. More... | |
| utils::Matrix< double > | node_histogram_distance (SampleSet const &sample_set, size_t const histogram_bins=25) |
| Calculate the Node Histogram Distance of every pair of Samples in the SampleSet. More... | |
| utils::Matrix< double > | node_histogram_distance (std::vector< NodeDistanceHistogramSet > const &histogram_sets) |
| Given the histogram sets that describe a set of Samples, calculate their pairwise distance matrix. More... | |
| void | normalize_weight_ratios (Pquery &pquery) |
Recalculate the like_weight_ratio of the PqueryPlacement&s of a Pquery, so that their sum is 1.0, while maintaining their ratio to each other. More... | |
| void | normalize_weight_ratios (Sample &smp) |
Recalculate the like_weight_ratio of the PqueryPlacement&s of each Pquery in the Sample, so that their sum is 1.0, while maintaining their ratio to each other. More... | |
| std::ostream & | operator<< (std::ostream &out, Sample const &smp) |
| Print a table of all Pqueries with their Placements and Names to the stream. More... | |
| std::ostream & | operator<< (std::ostream &out, SampleSet const &sample_set) |
| std::ostream & | operator<< (std::ostream &out, SimulatorEdgeDistribution const &distrib) |
| std::ostream & | operator<< (std::ostream &out, SimulatorExtraPlacementDistribution const &distrib) |
| std::ostream & | operator<< (std::ostream &out, SimulatorLikeWeightRatioDistribution const &distrib) |
| double | pairwise_distance (const Sample &smp_a, const Sample &smp_b, bool with_pendant_length=false) |
| Calculate the normalized pairwise distance between all placements of the two Samples. More... | |
| std::vector< utils::Color > | placement_color_count_gradient (Sample const &smp, bool linear) |
| Returns a vector with a Color for each edge that visualizes the number of placements on that edge. More... | |
| std::pair< PlacementTreeEdge const *, size_t > | placement_count_max_edge (Sample const &smp) |
| Get the number of placements on the edge with the most placements, and a pointer to this edge. More... | |
| std::vector< size_t > | placement_count_per_edge (Sample const &sample) |
| Return a vector that contains the number of PqueryPlacements per edge of the tree of the Sample. More... | |
| utils::Matrix< size_t > | placement_count_per_edge (SampleSet const &sample_set) |
| double | placement_distance (PqueryPlacement const &place_a, PqueryPlacement const &place_b, utils::Matrix< double > const &node_distances) |
| Calculate the distance between two PqueryPlacements, using their position on the tree::TreeEdges, measured in branch length units. More... | |
| double | placement_distance (PqueryPlacement const &placement, tree::TreeNode const &node, utils::Matrix< double > const &node_distances) |
| Calculate the distance in branch length units between a PqueryPlacement and a tree::TreeNode. More... | |
| std::pair< PlacementTreeEdge const *, double > | placement_mass_max_edge (Sample const &smp) |
Get the summed mass of the placements on the heaviest edge, measured by their like_weight_ratio, and a pointer to this edge. More... | |
| std::vector< double > | placement_mass_per_edge_without_multiplicities (Sample const &sample) |
| Return a vector that contains the sum of the masses of the PqueryPlacements per edge of the tree of the Sample. More... | |
| utils::Matrix< double > | placement_mass_per_edge_without_multiplicities (SampleSet const &sample_set) |
| Return a Matrix that contains the placement masses per edge. More... | |
| std::vector< double > | placement_mass_per_edges_with_multiplicities (Sample const &sample) |
| Return a vector that contains the sum of the masses of the PqueryPlacements per edge of the tree of the Sample, using the multiplicities as factors. More... | |
| utils::Matrix< double > | placement_mass_per_edges_with_multiplicities (SampleSet const &sample_set) |
| Return a Matrix that contains the placement masses per edge, using the multiplicities as factors. More... | |
| size_t | placement_path_length_distance (PqueryPlacement const &placement, tree::TreeEdge const &edge, utils::Matrix< size_t > const &edge_path_lengths) |
| Calculate the discrete distance from a PqueryPlacement to an edge, measured as the number of nodes between them. More... | |
| size_t | placement_path_length_distance (PqueryPlacement const &place_a, PqueryPlacement const &place_b, utils::Matrix< size_t > const &node_path_lengths) |
| std::vector< std::vector< PqueryPlacement const * > > | placements_per_edge (Sample const &smp, bool only_max_lwr_placements=false) |
| Return a mapping from each PlacementTreeEdges to the PqueryPlacements that are placed on that edge. More... | |
| std::vector< PqueryPlacement const * > | placements_per_edge (Sample const &smp, PlacementTreeEdge const &edge) |
| Return a vector of all PqueryPlacements that are placed on the given PlacementTreeEdge. More... | |
| std::vector< PqueryPlain > | plain_queries (Sample const &smp) |
| Return a plain representation of all pqueries of this map. More... | |
| std::vector< std::vector< Pquery const * > > | pqueries_per_edge (Sample const &sample, bool only_max_lwr_placements=false) |
| Return a mapping from each edge to the Pqueries on that edge. More... | |
| double | pquery_distance (Pquery const &pquery, tree::TreeNode const &node, utils::Matrix< double > const &node_distances) |
Calculate the weighted distance between the PqueryPlacements of a Pquery and a tree::TreeNode, in branch length units, using the like_weight_ratio of the PqueryPlacements for weighing. More... | |
| double | pquery_distance (Pquery const &pquery_a, Pquery const &pquery_b, utils::Matrix< double > const &node_distances, bool with_pendant_length=false) |
Calculate the weighted distance between two Pqueries, in branch length units, as the pairwise distance between their PqueryPlacements, and using the like_weight_ratio for weighing. More... | |
| template<typename DistanceFunction > | |
| double | pquery_distance (Pquery const &pquery, DistanceFunction distance_function) |
| Local helper function to avoid code duplication. More... | |
| template<typename DistanceFunction > | |
| double | pquery_distance (Pquery const &pquery_a, Pquery const &pquery_b, DistanceFunction distance_function) |
| Local helper function to avoid code duplication. More... | |
| double | pquery_distance (PqueryPlain const &pquery_a, PqueryPlain const &pquery_b, utils::Matrix< double > const &node_distances, bool with_pendant_length=false) |
| Calculate the weighted distance between two plain pqueries. It is mainly a helper method for distance calculations (e.g., pairwise distance, variance). More... | |
| double | pquery_path_length_distance (Pquery const &pquery, tree::TreeEdge const &edge, utils::Matrix< size_t > const &edge_path_lengths) |
Calculate the weighted discrete distance between the PqueryPlacements of a Pquery and a tree::TreeEdge, in number of nodes between them, using the like_weight_ratio of the PqueryPlacements for weighing. More... | |
| double | pquery_path_length_distance (Pquery const &pquery_a, Pquery const &pquery_b, utils::Matrix< size_t > const &node_path_lengths) |
Calculate the weighted discrete distance between two Pqueries, measured as the pairwise distance in number of nodes between between their PqueryPlacements, and using the like_weight_ratio for weighing. More... | |
| std::string | print_tree (Sample const &smp) |
| Return a simple view of the Tree of a Sample with information about the Pqueries on it. More... | |
| void | rectify_values (Sample &sample) |
| Correct invalid values of the PqueryPlacements and PqueryNames as good as possible. More... | |
| void | rectify_values (SampleSet &sset) |
| Correct invalid values of the PqueryPlacements and PqueryNames as good as possible. More... | |
| size_t | remove_empty_name_pqueries (Sample &sample) |
| Remove all Pqueries from the Sample that have no PqueryNames. More... | |
| size_t | remove_empty_placement_pqueries (Sample &sample) |
| Remove all Pqueries from the Sample that have no PqueryPlacements. More... | |
| void | reset_edge_nums (PlacementTree &tree) |
| Reset all edge nums of a PlacementTree. More... | |
| void | scale_all_branch_lengths (Sample &smp, double factor=1.0) |
| Scale all branch lengths of the Tree and the position of the PqueryPlacements by a given factor. More... | |
| void | set_depths_distributed_weights (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of an SimulatorEdgeDistribution so that they follow the depth distribution of the edges in the provided Sample. More... | |
| void | set_depths_distributed_weights (Sample const &sample, std::vector< double > const &depth_weights, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights so that they follow a given depth distribution of the edges in the PlacementTree. More... | |
| void | set_random_edges (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of a SimulatorEdgeDistribution randomly to either 0.0 or 1.0, so that a random subset of edges is selected (with the same probability for each selected edge). More... | |
| void | set_random_edges (size_t edge_count, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of an SimulatorEdgeDistribution randomly to either 0.0 or 1.0, so that a random subset of edges is selected (with the same probability for each selected edge). More... | |
| size_t | set_random_subtree_weights (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Sets the weights of an SimulatorEdgeDistribution to 1.0 for a randomly chosen subtree, all others to 0.0. More... | |
| void | set_random_weights (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of an SimulatorEdgeDistribution for the edges randomly to a value between 0.0 and 1.0. More... | |
| void | set_random_weights (size_t edge_count, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of an SimulatorEdgeDistribution for the edges randomly to a value between 0.0 and 1.0. More... | |
| void | set_subtree_weights (Sample const &sample, size_t link_index, SimulatorEdgeDistribution &edge_distrib) |
| Set the weights of a subtree to 1.0 and all other weights to 0.0. More... | |
| void | set_uniform_weights (Sample const &sample, SimulatorEdgeDistribution &edge_distrib) |
| Sets the weights of an SimulatorEdgeDistribution to 1.0 for all edges, so that each edge has the same probability of being chosen. More... | |
| void | set_uniform_weights (size_t edge_count, SimulatorEdgeDistribution &edge_distrib) |
| Sets the weights of an SimulatorEdgeDistribution to 1.0 for all edges, so that each edge has the same probability of being chosen. More... | |
| void | sort_placements_by_weight (Pquery &pquery) |
Sort the PqueryPlacements of a Pquery by their like_weight_ratio, in descending order (most likely first). More... | |
| void | sort_placements_by_weight (Sample &smp) |
Sort the PqueryPlacements of all Pqueries by their like_weight_ratio, in descending order (most likely first). More... | |
| void | swap (Sample &lhs, Sample &rhs) |
| double | total_multiplicity (Pquery const &pqry) |
| Return the sum of all multiplicities of the Pquery. More... | |
| double | total_multiplicity (Sample const &sample) |
| Return the sum of all multiplicities of all the Pqueries of the Sample. More... | |
| size_t | total_name_count (Sample const &smp) |
| Get the total number of PqueryNames in all Pqueries of the given Sample. More... | |
| size_t | total_placement_count (Sample const &smp) |
| Get the total number of PqueryPlacements in all Pqueries of the given Sample. More... | |
| double | total_placement_mass_with_multiplicities (Sample const &smp) |
| Get the mass of all PqueryPlacements of the Sample, using the multiplicities as factors. More... | |
| double | total_placement_mass_without_multiplicities (Sample const &smp) |
| Get the summed mass of all PqueryPlacements in all Pqueries of the given Sample, where mass is measured by the like_weight_ratios of the PqueryPlacements. More... | |
| size_t | total_pquery_count (SampleSet const &sample_set) |
| Return the total number of Pqueries in the Samples of the SampleSet. More... | |
| tree::TreeSet | tree_set (SampleSet const &sample_set) |
| Return a TreeSet containing all the trees of the SampleSet. More... | |
| bool | validate (Sample const &smp, bool check_values=false, bool break_on_values=false) |
| Validate the integrity of the pointers, references and data in a Sample object. More... | |
| double | variance (const Sample &smp, bool with_pendant_length=false) |
| Calculate the variance of the placements on a tree. More... | |
| static double | variance_partial_ (PqueryPlain const &pqry_a, std::vector< PqueryPlain > const &pqrys_b, utils::Matrix< double > const &node_distances, bool with_pendant_length) |
| Internal function that calculates the sum of distances contributed by one pquery for the variance. See variance() for more information. More... | |
Typedefs | |
| using | PlacementTree = tree::Tree |
| Alias for a tree::Tree used for a tree with information needed for storing Pqueries. This kind of tree is used by Sample. More... | |
| using | PlacementTreeEdge = tree::TreeEdge |
| Alias for tree::TreeEdge used in a PlacementTree. See PlacementEdgeData for the data stored on the edges. More... | |
| using | PlacementTreeLink = tree::TreeLink |
| Alias for tree::TreeLink used in a PlacementTree. More... | |
| using | PlacementTreeNode = tree::TreeNode |
| Alias for tree::TreeNode used in a PlacementTree. See PlacementNodeData for the data stored on the nodes. More... | |
| double add_sample_to_mass_tree | ( | Sample const & | smp, |
| double const | sign, | ||
| double const | scaler, | ||
| tree::MassTree & | target | ||
| ) |
Helper function to copy masses from a Sample to a MassTree.
The function copies the masses from a Sample to a MassTree. It returns the amount of work needed to move the masses from their pendant position to the branch (this result is only used if with_pendant_length is true in the calculation functions).
Definition at line 132 of file placement/function/operators.cpp.
| void adjust_branch_lengths | ( | Sample & | sample, |
| tree::Tree const & | source | ||
| ) |
Take the branch lengths of the source Tree and use them as the new branch lengths of the sample.
The proximal_lengths of the PqueryPlacements are adjusted accordingly, so that their relative position on the branch stays the same.
The source Tree is expected to have edges with data type tree::CommonEdgeData.
The topology of the source and the tree of the Sample have to be identical. This is however not checked, so the user has to provide a fitting tree.
Definition at line 178 of file placement/function/functions.cpp.
| void adjust_branch_lengths | ( | SampleSet & | sample_set, |
| tree::Tree const & | source | ||
| ) |
Take the branch lengths of the source Tree and use them as the new branch lengths of the Samples in the sample_set.
This function simply calls adjust_branch_lengths( Sample&, tree::Tree const& ) for all Samples in the set. See there for details.
All involved Trees need to have identical topology. This is not checked.
Definition at line 165 of file sample_set.cpp.
| void adjust_to_average_branch_lengths | ( | SampleSet & | sample_set | ) |
Set the branch lengths of all Samples in the sample_set to the respecitve average branch length of the Samples.
That is, for each edge of the tree, find the average branch length over all Samples, and use this for the Samples. This means, all Samples in the SampleSet need to have identical tree topologies.
Definition at line 172 of file sample_set.cpp.
| bool all_identical_trees | ( | SampleSet const & | sample_set | ) |
Returns true iff all Trees of the Samples in the set are identical.
This is the case if they have the same topology, node names and edge_nums. However, branch lengths are not checked, because usually those differ slightly.
Definition at line 125 of file sample_set.cpp.
| std::unordered_set< std::string > all_pquery_names | ( | Sample const & | sample | ) |
Return a set of all unique PqueryNames of the Pqueries of the given sample.
If a Pquery contains multiple names, all of them are added to the set.
Definition at line 108 of file placement/function/functions.cpp.
| tree::Tree average_branch_length_tree | ( | SampleSet const & | sample_set | ) |
Return the Tree that has edges with the average branch length of the respective edges of the Trees in the Samples of the given SampleSet.
Definition at line 120 of file sample_set.cpp.
| std::pair< PlacementTreeEdge const *, double > center_of_gravity | ( | Sample const & | smp, |
| bool const | with_pendant_length = false |
||
| ) |
Calculate the Center of Gravity of the placements on a tree.
The center of gravity is the point on the tree where all masses of the placements on the one side of it times their distance from the point are equal to this sum on the other side of the point. In the following example, the hat ^ marks this point on a line with two placements: One has mass 1 and distance 3 from the central point, and one as mass 3 and distance 1, so that the product of their mass and distance to the point is the same:
3
|
1 |
|_____________|
^
It is thus like calculating masses and torques on a lever in order to find their physical center of mass/gravity.
This calculation is done for the whole tree, with the masses calculated from the like_weight_ratio and distances in terms of the branch_length of the edges and the proximal_length and (if specificed in the method parameter) the pendant_length of the placements.
| double center_of_gravity_distance | ( | Sample const & | smp_a, |
| Sample const & | smp_b, | ||
| bool const | with_pendant_length = false |
||
| ) |
Calculate the distance between the two Centers of Gravity of two Samples.
The distance is measured in branch length units; for the Center of Gravity, see center_of_gravity().
| double center_of_gravity_variance | ( | Sample const & | smp, |
| bool const | with_pendant_length = false |
||
| ) |
Calculate the variance of the PqueryPlacements of a Sample around its Center of Gravity.
The caluclation of the variance is as follows:
\( Var(X) = E[ (x - \mu)^2 ] = \frac{\sum (x - \mu)^2 \cdot \omega} {\sum \omega} \), where the weights \( \omega \) are the like_weight_ratios of the placements.
See center_of_gravity() for more.
| std::vector< int > closest_leaf_depth_histogram | ( | Sample const & | smp | ) |
Return a histogram representing how many placements have which depth with respect to their closest leaf node.
The depth between two nodes on a tree is the number of edges between them. Thus, the depth of a placement (which sits on an edge of the tree) to a specific node is the number of edges between this node and the closer one of the two nodes at the end of the edge where the placement sits.
The closest leaf to a placement is thus the leaf node which has the smallest depth to that placement. This function then returns a histogram of how many placements (values of the vector) are there that have a specific depth (indices of the vector) to their closest leaf.
Example: A return vector of
histogram[0] = 2334 histogram[1] = 349 histogram[2] = 65 histogram[3] = 17
means that there are 2334 placements that sit on an edge which leads to a leaf node (thus, the depth of one of the nodes of the edge is 0). It has 349 placements that sit on an edge where one of its nodes has one neighbour that is a leaf; and so on.
The vector is automatically resized to the needed number of elements.
Definition at line 858 of file placement/function/functions.cpp.
| std::vector< int > closest_leaf_distance_histogram | ( | Sample const & | smp, |
| const double | min, | ||
| const double | max, | ||
| const int | bins = 10 |
||
| ) |
Returns a histogram counting the number of placements that have a certain distance to their closest leaf node, divided into equally large intervals between a min and a max distance.
The distance range between min and max is divided into bins many intervals of equal size. Then, the distance from each placement to its closest leaf node is calculated and the counter for this particular distance inverval in the histogram is incremented.
The distance is measured along the branch_length values of the edges, taking the pendant_length and proximal_length of the placements into account. If the distances is outside of the interval [min,max], the counter of the first/last bin is incremented respectively.
Example:
double min = 0.0;
double max = 20.0;
int bins = 25;
double bin_size = (max - min) / bins;
std::vector<int> hist = closest_leaf_distance_histogram (min, max, bins);
for (unsigned int bin = 0; bin < hist.size(); ++bin) {
LOG_INFO << "Bin " << bin << " [" << bin * bin_size << "; "
<< (bin+1) * bin_size << ") has " << hist[bin] << " placements.";
}
%
Definition at line 884 of file placement/function/functions.cpp.
| std::vector< int > closest_leaf_distance_histogram_auto | ( | Sample const & | smp, |
| double & | min, | ||
| double & | max, | ||
| const int | bins = 10 |
||
| ) |
Returns the same type of histogram as closest_leaf_distance_histogram(), but automatically determines the needed boundaries.
See closest_leaf_distance_histogram() for general information about what this function does. The difference between both functions is that this one first procresses all distances from placements to their closest leaf nodes to find out what the shortest and longest are, then sets the boundaries of the histogram accordingly. The number of bins is then used to divide this range into intervals of equal size.
The boundaries are returned by passing two doubles min and max to the function by reference. The value of max will actually contain the result of std::nextafter() called on the longest distance; this makes sure that the value itself will be placed in the interval.
Example:
double min, max;
int bins = 25;
std::vector<int> hist = closest_leaf_distance_histogram (min, max, bins);
double bin_size = (max - min) / bins;
LOG_INFO << "Histogram boundaries: [" << min << "," << max << ").";
for (unsigned int bin = 0; bin < hist.size(); ++bin) {
LOG_INFO << "Bin " << bin << " [" << bin * bin_size << "; "
<< (bin+1) * bin_size << ") has " << hist[bin] << " placements.";
}
It has a slightly higher time and memory consumption than the non-automatic version closest_leaf_distance_histogram(), as it needs to process the values twice in order to find their min and max.
Definition at line 922 of file placement/function/functions.cpp.
| std::vector< double > closest_leaf_weight_distribution | ( | Sample const & | sample | ) |
Definition at line 830 of file placement/function/functions.cpp.
| void collect_duplicate_pqueries | ( | Sample & | smp | ) |
Find all Pqueries that share a common name and combine them into a single Pquery containing all their collective PqueryPlacements and PqueryNames.
The function collects all Pqueries that share at least one name. This is transitive, so that for example three Pqueries with two names each like (a,b) (b,c) (c,d) will be combined into one Pquery. Thus, the transitive closure of shared names is collected.
All those Pqueries with shared names are combined by simply moving all their Placements and Names into one Pquery and deleting the others. This means that at least the shared names will be doubled after this function. Also, Placements on the same edge can occur. Thus, usually merge_duplicate_names() and merge_duplicate_placements() are called after this function. The function merge_duplicates() does exaclty this, for convenience.
Definition at line 576 of file placement/function/functions.cpp.
| bool compatible_trees | ( | PlacementTree const & | lhs, |
| PlacementTree const & | rhs | ||
| ) |
Return whether two PlacementTrees are compatible.
This is the case iff:
In all other cases, false is returned.
Definition at line 63 of file placement/function/operators.cpp.
Return whether the PlacementTrees of two Samples are compatible.
See this version of the function for details.
Definition at line 96 of file placement/function/operators.cpp.
| PlacementTree convert_common_tree_to_placement_tree | ( | tree::CommonTree const & | source_tree | ) |
Convert a CommonTree into a PlacementTree.
This function returns a new tree with the same topology as the source tree, and the same node names and branch lengths. In addition, the edge_num property of the PlacementTree is established, as it is not part of the CommonTree data.
Definition at line 105 of file placement/function/operators.cpp.
| std::pair< tree::TreeSet, std::vector< double > > convert_sample_set_to_mass_trees | ( | SampleSet const & | sample_set, |
| bool | normalize | ||
| ) |
Convert all Samples in a SampleSet to tree::MassTrees.
Definition at line 185 of file placement/function/operators.cpp.
| std::pair< tree::MassTree, double > convert_sample_to_mass_tree | ( | Sample const & | sample, |
| bool | normalize | ||
| ) |
Convert a Sample to a tree::MassTree.
The function takes all PqueryPlacements of the Sample and adds their masses in form of the like_weight_ratio as mass points on a tree::MassTree.
Definition at line 174 of file placement/function/operators.cpp.
Copy all Pqueries from the source Sample (left parameter) to the target Sample (right parameter).
For this method to succeed, the PlacementTrees of the Samples need to have the same topology, including identical edge_nums and node names. Otherwise, this function throws an std::runtime_error.
The PlacementTree of the target Sample is not modified. If the average branch length tree is needed instead, see SampleSet::merge_all().
Definition at line 539 of file placement/function/functions.cpp.
| double earth_movers_distance | ( | Sample const & | lhs, |
| Sample const & | rhs, | ||
| double const | p = 1.0, |
||
| bool const | with_pendant_length = false |
||
| ) |
Calculate the earth mover's distance between two Samples.
This function interprets the like_weight_ratios of the PqueryPlacements as masses distributed along the branches of a tree. It then calculates the earth mover's distance between those masses for the distrubitons induced by the two given Samples.
In order to do so, first, a tree with the average branch lengths of the two PlacementTrees is calculated. This is because of numerical issues that might yield different branch lengths. This necessiates that the trees have the same topology. If not, an std::runtime_error is thrown. The masses are then distributed on this tree, using the same relative position on their branches that they had in their original trees.
The calculation furthermore takes the multiplicities of the Pqueries into account. That means, pqueries with higher (total) multiplicity have a higher influence on the calculated distance.
As the two Samples might have a different total number of Pqueries, the masses of the Samples are first normalized to 1.0, using all the like_weight_ratios and multiplicities of the Pqueries. As a consequence, the resulting distance will not reflect the total number of Pqueries, but only their relative (normalized) distrubution on the tree.
Furthermore, the parameter p is used to control the influence of mass and distance, with 0.0 < p < inf, and default p == 1.0, which is the neutral case. A larger p increases the impact of distance traveled, while a smaller p emphasizes differences of mass.
See earth_movers_distance( MassTree const&, MassTree const& ) for more information on the actual distance calculation and details on the parameter p.
Definition at line 67 of file placement/function/emd.cpp.
| utils::Matrix< double > earth_movers_distance | ( | SampleSet const & | sample_set, |
| double const | p = 1.0, |
||
| bool const | with_pendant_length = false |
||
| ) |
Calculate the pairwise Earth Movers Distance for all Samples in a SampleSet.
The result is a pairwise distance Matrix using the indices of the Samples in the SampleSet. See earth_movers_distance( Sample const&, Sample const&, ... ) for details on this distance measure on Samples, and see earth_movers_distance( MassTree const&, MassTree const& ) for more information on the actual distance calculation, and the parameter p.
Definition at line 107 of file placement/function/emd.cpp.
| std::unordered_map< int, PlacementTreeEdge * > edge_num_to_edge_map | ( | PlacementTree const & | tree | ) |
Return a mapping of edge_num integers to the corresponding PlacementTreeEdge object.
In a valid jplace file, the edge_nums are in increasing order with a postorder traversal of the tree. However, as Genesis does not need this constraint, we return a map here instead.
Definition at line 55 of file placement/function/helper.cpp.
| std::unordered_map< int, PlacementTreeEdge * > edge_num_to_edge_map | ( | Sample const & | smp | ) |
Return a mapping of edge_num integers to the corresponding PlacementTreeEdge object.
This function depends on the tree only and does not involve any pqueries. Thus, it forwards to edge_num_to_edge_map( PlacementTree const& ). See there for details.
Definition at line 66 of file placement/function/helper.cpp.
| double edpl | ( | Pquery const & | pquery, |
| utils::Matrix< double > const & | node_distances | ||
| ) |
Calculate the EDPL uncertainty values for a Pquery.
This is the function that does the actual computation. It is used by the other edpl functions, which first calculate the node_distances matrix before calling this function. It is useful to separate these steps in order to avoid duplicate work when calculating the edpl for many Pqueries at a time.
node_distances has to be the result of node_branch_length_distance_matrix().
Definition at line 75 of file measures.cpp.
| std::vector< double > edpl | ( | Sample const & | sample | ) |
Calculate the expected distance between placement locations (EDPL) for all Pqueries in a Sample.
The EDPL is a measure of uncertainty of how far the PqueryPlacements of a Pquery are spread across the branches of the PlacementTree. In a reference tree with similar sequences, a query sequence might be placed on several nearby branches with relatively high likelihood (LWR). This still constitutes a high confidence in the placement, as the spreading is due to the similar reference sequences, and not due to inherent uncertainty in the placement itself. This is opposed to a query sequence whose placements are spread all across the tree, which might indicate that a fitting reference sequence is missing from the tree, and hence yields uncertain placements.
This can be assessed with the EDPL, which calculates the distances between different placements, weighted by their respective LWRs:
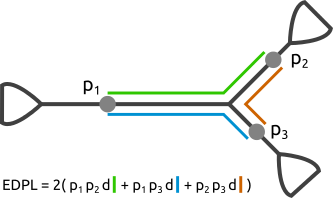
The p values in the figure represent likelihood weight ratios of the placements at these locations. The distances d are calculated using the branch lengths of the tree on the path between the placement locations. Hence, a low EDPL indicates that the PqueryPlacements of a Pquery are focussed in a narrow region of the tree, whereas a high EDPL indicates that the placements are spread across the tree.
See http://matsen.github.io/pplacer/generated_rst/guppy_edpl.html for more information. The function calculates the node distances of the tree first, which is needed for the computation. See the other edpl functions for versions that take this matrix, in order to get speedups when working with multiple Samples that use the same PlacementTree.
Definition at line 117 of file measures.cpp.
Calculate the EDPL uncertainty values for a Pquery.
See http://matsen.github.io/pplacer/generated_rst/guppy_edpl.html for more information.
This function expects a Pquery and the Sample it belongs to. This is necessary in order to get the Tree of the Sample and calculate distances between its Nodes.
Definition at line 111 of file measures.cpp.
| std::vector< double > edpl | ( | Sample const & | sample, |
| utils::Matrix< double > const & | node_distances | ||
| ) |
Calculate the edpl() for all Pqueries in the Sample.
node_distances has to be the result of node_branch_length_distance_matrix().
Definition at line 97 of file measures.cpp.
| EpcaData epca | ( | SampleSet const & | samples, |
| double | kappa = 1.0, |
||
| double | epsilon = 1e-5, |
||
| size_t | components = 0 |
||
| ) |
Perform EdgePCA on a SampleSet.
The parameters kappa and epsilon are as described in epca_splitify_transform() and epca_filter_constant_columns(), respectively.
The result is returned as a struct similar to the one used by utils::pca(), but containing an additional vector of the edge indices that the rows of the eigenvectors Matrix correspond to. This is necessary for back-mapping the eigenvectors onto the edges of the tree.
| utils::Matrix< double > epca_imbalance_matrix | ( | SampleSet const & | samples, |
| bool | include_leaves = false, |
||
| bool | normalize = true |
||
| ) |
Calculate the imbalance matrix of placment mass for all Samples in a SampleSet.
The first step to perform Edge PCA is to make a Matrix with rows indexed by the Samples, and columns by the Edges of the Tree. Each entry of this matrix is the difference between the distribution of mass on either side of an edge for a Sample. Specifically, it is the amount of mass on the distal (non-root) side of the edge minus the amount of mass on the proximal side.
The matrix is row-indexed according to the Samples in the SampleSet.
If include_leaves is set to false (default), the columns for edges belonging to leaves of the tree are left out. Their value is -1.0 anyway, as there is no mass on the distal side of those edges. Hence, they are constant for all Samples and have no effect on the Edge PCA result. In this case, the matrix is column-indexed so that each inner edge of the Tree has one column in the Matrix. See epca_imbalance_vector() for more details.
If include_leaves is set to true, the constant values for leaf edges are also included. In this case, the matrix is column-indexed according to the edge indices of the Tree. This is for example useful if the indexing is needed later. The columns can then also be filtered out using epca_filter_constant_columns().
Lastly normalize is used as in epca_imbalance_vector(). See there for details.
| std::vector< double > epca_imbalance_vector | ( | Sample const & | sample, |
| bool | normalize = true |
||
| ) |
Calculate the imbalance of placement mass for each Edge of the given Sample.
The entries of the vector are the difference between the distribution of mass on either side of the edge for the given Sample. Specifically, it is the amount of mass on the distal (non-root) side of the edge minus the amount of mass on the proximal (root) side.
If normalize is true (default), the imbalance values are normalized by the total amount of mass on the tree (expect for the mass of the respective edge, as this one also does not count for its own imbalance).
The vector is indexed using the index() of the edges. This is different from how how guppy indexes the edges, namely by using their edge_nums. See https://matsen.github.io/pplacer/generated_rst/guppy_splitify.html for details on the guppy edge imbalance matrix. We chose to use our internal edge index instead, as it is consistent and needs no checking for correctly labeled edge nums.
| void epca_splitify_transform | ( | utils::Matrix< double > & | imbalance_matrix, |
| double | kappa = 1.0 |
||
| ) |
Perform a component-wise transformation of the imbalance matrix used for epca().
All entries of the Matrix are transformed inplace, using
\[ \varphi_\kappa(x) = \mathrm{sgn}(x) \cdot |x|^\kappa \]
where the kappa ( \(\kappa\)) parameter can be any non-negative number. This parameter scales between ignoring abundance information (kappa = 0), using it linearly (kappa = 1), and emphasizing it (kappa > 1).
| [in,out] | imbalance_matrix | Matrix to transform inplace. |
| [in] | kappa | Scaling value for abundance information. Has to be >= 0. |
|
static |
| void filter_max_pendant_length | ( | Pquery & | pquery, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a pendant_length above the given threshold.
Definition at line 318 of file placement/function/functions.cpp.
| void filter_max_pendant_length | ( | Sample & | sample, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a pendant_length above the given threshold from all Pqueries of the Sample.
Definition at line 326 of file placement/function/functions.cpp.
| void filter_min_accumulated_weight | ( | Pquery & | pquery, |
| double | threshold = 0.99 |
||
| ) |
Remove the PqueryPlacements with the lowest like_weight_ratio, while keeping the accumulated weight (sum of all remaining like_weight_ratios) above a given threshold.
This is a cleaning function to get rid of unlikely placement positions, withouth sacrificing too much detail of the overall distribution of weights. The EPA support a similar option, which only writes enough of the most likely placement positions to the output to fullfil a threshold.
Definition at line 212 of file placement/function/functions.cpp.
| void filter_min_accumulated_weight | ( | Sample & | smp, |
| double | threshold = 0.99 |
||
| ) |
Remove the PqueryPlacements with the lowest like_weight_ratio, while keeping the accumulated weight (sum of all remaining like_weight_ratios) above a given threshold.
This function calls filter_min_accumulated_weight( Pquery& pquery, double threshold ) for all Pqueries of the Sample. See this version of the function for more information.
Definition at line 239 of file placement/function/functions.cpp.
| void filter_min_pendant_length | ( | Pquery & | pquery, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a pendant_length below the given threshold.
Definition at line 303 of file placement/function/functions.cpp.
| void filter_min_pendant_length | ( | Sample & | sample, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a pendant_length below the given threshold from all Pqueries of the Sample.
Definition at line 311 of file placement/function/functions.cpp.
| void filter_min_weight_threshold | ( | Pquery & | pquery, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a like_weight_ratio below the given threshold.
Definition at line 275 of file placement/function/functions.cpp.
| void filter_min_weight_threshold | ( | Sample & | smp, |
| double | threshold | ||
| ) |
Remove all PqueryPlacements that have a like_weight_ratio below the given threshold from all Pqueries of the Sample.
Definition at line 296 of file placement/function/functions.cpp.
| void filter_n_max_weight_placements | ( | Pquery & | pquery, |
| size_t | n = 1 |
||
| ) |
Remove all PqueryPlacements but the n most likely ones from the Pquery.
Pqueries can contain multiple placements on different branches. For example, the EPA algorithm of RAxML outputs up to the 7 most likely positions for placements to the output Jplace file by default. The property like_weight_ratio weights those placement positions so that the sum over all positions (all branches of the tree) per pquery is 1.0.
This function removes all but the n most likely placements (the ones which have the highest like_weight_ratio) from the Pquery. The like_weight_ratio of the remaining placements is not changed.
Definition at line 246 of file placement/function/functions.cpp.
| void filter_n_max_weight_placements | ( | Sample & | smp, |
| size_t | n = 1 |
||
| ) |
Remove all PqueryPlacements but the n most likely ones from all Pqueries in the Sample.
This function calls filter_n_max_weight_placements( Pquery& pquery, size_t n ) for all Pqueries of the Sample. See this version of the function for more information.
Definition at line 268 of file placement/function/functions.cpp.
| void genesis::placement::filter_pqueries_by_name_ | ( | Sample & | smp, |
| F | predicate, | ||
| bool | remove_empty_pqueries | ||
| ) |
Definition at line 396 of file placement/function/functions.cpp.
| void filter_pqueries_differing_names | ( | Sample & | sample_1, |
| Sample & | sample_2, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames from the two Samples that occur in both of them.
This function builds the intersection of the set of names of both Samples and removes all those PqueryNames that occur in both sets.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 474 of file placement/function/functions.cpp.
| void filter_pqueries_intersecting_names | ( | Sample & | sample_1, |
| Sample & | sample_2, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames from the two Samples that are unique to each of them.
This function builds the intersection of the set of names of both Samples and only keeps those PqueryNames that occur in both sets.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 460 of file placement/function/functions.cpp.
| void filter_pqueries_keeping_names | ( | Sample & | smp, |
| std::string const & | regex, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames which do not match the given regex.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 418 of file placement/function/functions.cpp.
| void filter_pqueries_keeping_names | ( | Sample & | smp, |
| std::unordered_set< std::string > const & | keep_list, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames which do not occur in the given keep_list.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 429 of file placement/function/functions.cpp.
| void filter_pqueries_removing_names | ( | Sample & | smp, |
| std::string const & | regex, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames which match the given regex.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 439 of file placement/function/functions.cpp.
| void filter_pqueries_removing_names | ( | Sample & | smp, |
| std::unordered_set< std::string > const & | remove_list, | ||
| bool | remove_empty_name_pqueries = true |
||
| ) |
Remove all PqueryNames which occur in the given remove_list.
If all names from a given Pquery are removed that way, and if the remove_empty_name_pqueries flag is true (default), the whole Pquery is removed from the Sample as well.
Definition at line 450 of file placement/function/functions.cpp.
Return the first Pquery that has a particular name, or nullptr of none has.
Definition at line 96 of file placement/function/functions.cpp.
Return the first Pquery that has a particular name, or nullptr of none has.
Definition at line 84 of file placement/function/functions.cpp.
Get the first Sample in a SampleSet that has a given name, or nullptr if not found.
Definition at line 48 of file sample_set.cpp.
Get the first Sample in a SampleSet that has a given name, or nullptr if not found.
Definition at line 58 of file sample_set.cpp.
| bool has_consecutive_edge_nums | ( | PlacementTree const & | tree | ) |
Verify that the PlacementTree has no duplicate edge_nums and that they form consecutive numbers starting from 0.
This function is very similar to has_correct_edge_nums(). However, instead of checking whether the edge_nums are correctly assigned following a postorder traversal of the tree, as demanded by the Jplace standard, this function simply checks whehter they are all unique, start at 0 and continue consecutively without gaps.
This is imporant for using the edge_nums as indices, for example.
We offer this function, because Genesis can work with improperly assigned edge_nums, but for some functions we need to ensure those properties. Generally, you should however prefer correct edge_nums according to the standard, and use has_correct_edge_nums() to verify them.
Definition at line 300 of file placement/function/helper.cpp.
| bool has_correct_edge_nums | ( | PlacementTree const & | tree | ) |
Verify that the tree has correctly set edge nums.
The edge_num property of the PlacementTreeEdges is defined by the jplace standard. The values have to be assigned increasingly with a postorder traversal of the tree. This function checks whether this is the case.
See also has_consecutive_edge_nums() for a relaxed version of this function, which might also be useful in some cases where the strict correct order according to the standard is not needed.
Definition at line 323 of file placement/function/helper.cpp.
| bool has_name | ( | Pquery const & | pquery, |
| std::string const & | name | ||
| ) |
Return true iff the given Pquery contains a particular name.
Definition at line 64 of file placement/function/functions.cpp.
| bool has_name | ( | Sample const & | smp, |
| std::string const & | name | ||
| ) |
Return true iff the given Sample contains a Pquery with a particular name, i.e., a PqueryName whose name member equals the given name.
Definition at line 74 of file placement/function/functions.cpp.
| tree::Tree labelled_tree | ( | Sample const & | sample, |
| bool | fully_resolve = false, |
||
| std::string const & | name_prefix = "" |
||
| ) |
Produce a Tree where the most probable PqueryPlacement of each Pquery in a Sample is turned into an Edge.
The function takes the original Tree of the Sample, and for each Pquery of the Sample, it attaches a new leaf Node to it. The new leaf represents the most probable PqueryPlacement of the Pquery, measured using the like_weight_ratio. The leaf is positioned according to the proximal_length and pendant_length of the PqueryPlacement. The resulting Tree is useful to get an overview of the distribution of placements. It is mainly intended to view a few placements. For large Samples, it might be a bit cluttered.
Similar trees are produced by RAxML EPA, where the file is called RAxML_labelledTree, and by the pplacer guppy tog command. Both programs differ in the exact way the the placements are added as edges. To control this behaviour, use the fully_resolve parameter.
Parameter fully_resolve == false
If fully_resolve is set to false (default), all placements at one edge are collected as children of one central base edge:
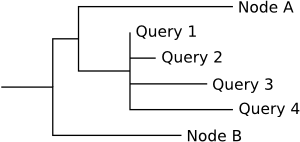
This method is similar to the way RAxML produces a labelled tree.
The base edge is positioned on the original edge at the average proximal_length of the placements. The base edge has a multifurcation if there are more than two placements on the edge.
The pendant_length of the placements is used to calculate the branch_length of the new placement edges. This calculation subtracts the shortest pendant_length of the placements on the edge, so that the base edge is maximally "moved" towards the placement edges. This also implies that at least one of the placement edges has branch_length == 0.0. Furthermore, the placements are sorted by their pendant_length.
Using this method, the new nodes of the resulting tree are easier to distinguish and collapse, as all placements are collected under the base edge. However, this comes at the cost of losing the detailled information of the proximal_length of the placements. If you want to keep this information, use fully_resolve == true instead.
Parameter fully_resolve == true
If fully_resolve is set to true, the placements are turned into single leaf nodes:
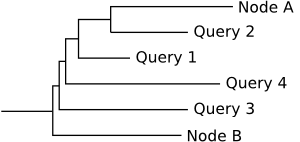
This method is similar to the way guppy tog produces a labelled tree.
The original edge is splitted into separate parts where each placement edge is attached. The branch_lengths between those parts are calculated using the proximal_length of the placements, while the branch_lengths of the placement edges use their pendant_length.
Using this method gives maximum information, but results in a more crowded tree. The new placement edges are "sorted" along the original edge by their proximal_length. For this reason in the example image above, "Query 2" is closer to "Node A" then "Query 1": it has a higher proximal_length. This information was lost in the multifurcating tree from above.
Further Details
For edges that contain only a single placement, both versions of fully_resolve behave the same. In this case, the placement is simply attached using its proximal_length and pendant_length.
Pqueries with multiple PqueryNames are treated as if each name is a separate placement, i.e., for each of them, a new (identical) edge is added to the Tree. If using fully_resolve == true, this results in a branch_length of 0.0 between the nodes of those placements.
| sample | Input Sample to get the Tree and PqueryPlacements from. |
| fully_resolve | Control in which way multiple placements at one edge are turned into new edges. See above for details. |
| name_prefix | Specify a prefix to be added to all new leaf Nodes (the ones that represent Placements). This is useful if a PqueryName also occurs as a name in the original tree. By default, empty. In order to get the same naming as labelled trees as produced by RAxML, use QUERY___. |
Definition at line 337 of file placement/function/tree.cpp.
| tree::Tree labelled_tree | ( | Sample const & | sample, |
| tree::Tree const & | tree, | ||
| bool | fully_resolve = false, |
||
| std::string const & | name_prefix = "" |
||
| ) |
Produce a Tree where each PqueryPlacement of a Sample is turned into an Edge.
This function is an extension of labelled_tree( Sample const&, bool, std::string const& ) that takes a custom Tree instead of using the one of the Sample. This allows to produce a labelled Tree that can contain other data at its Nodes and Edges. This Tree has to be topologically identical to the Sample Tree.
Furthermore, the data of the provided Tree needs to derived from CommonNodeData and CommonEdgeData. This data is then copied to the resulting Tree. The edge data of edges where new placement edges are added is kept that the topmost edge, i.e., the one that is closest to the root.
Definition at line 351 of file placement/function/tree.cpp.
| void genesis::placement::labelled_tree_add_lonely_placement_ | ( | tree::Tree & | tree, |
| tree::TreeEdge & | edge, | ||
| LabelledTreePlacementPair const & | placement_pair, | ||
| std::string const & | name_prefix | ||
| ) |
Definition at line 117 of file placement/function/tree.cpp.
| std::vector< std::vector< LabelledTreePlacementPair > > genesis::placement::labelled_tree_placement_pairs_per_edge_ | ( | Sample const & | sample | ) |
Definition at line 74 of file placement/function/tree.cpp.
| void genesis::placement::labelled_tree_process_edge_fully_resolved_ | ( | tree::Tree & | tree, |
| tree::TreeEdge & | edge, | ||
| std::vector< LabelledTreePlacementPair > const & | placement_pairs, | ||
| std::string const & | name_prefix | ||
| ) |
Definition at line 162 of file placement/function/tree.cpp.
| void genesis::placement::labelled_tree_process_edge_multifurcating_ | ( | tree::Tree & | tree, |
| tree::TreeEdge & | edge, | ||
| std::vector< LabelledTreePlacementPair > const & | placement_pairs, | ||
| std::string const & | name_prefix | ||
| ) |
Definition at line 245 of file placement/function/tree.cpp.
| void learn_like_weight_ratio_distribution | ( | Sample const & | sample, |
| SimulatorLikeWeightRatioDistribution & | lwr_distib, | ||
| size_t | number_of_intervals | ||
| ) |
Definition at line 423 of file placement/simulator/functions.cpp.
| void learn_per_edge_weights | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Sets the weights of an SimulatorEdgeDistributionso that they follow the same distribution of placement weight per edge as a given Sample.
This method "learns" how the placements on the given Sample are distributed by summing up their weight per edge and using this as weights. This way, the given distribution can be imitated by randomly generated placements.
The method is intended to be used on a Tree that has the same topology as the one that is given with the Sample, otherwise the Edge indices will not fit.
Definition at line 355 of file placement/simulator/functions.cpp.
| void learn_placement_number_weights | ( | Sample const & | sample, |
| SimulatorExtraPlacementDistribution & | p_distib | ||
| ) |
Definition at line 364 of file placement/simulator/functions.cpp.
| void learn_placement_path_length_weights | ( | Sample const & | sample, |
| SimulatorExtraPlacementDistribution & | p_distib | ||
| ) |
Definition at line 379 of file placement/simulator/functions.cpp.
|
static |
| void make_rooted | ( | Sample & | sample, |
| PlacementTreeEdge & | target_edge | ||
| ) |
Root the underlying PlacementTree of a Sample at a specified TreeEdge.
The function inserts a new root node on the given target_edge, which splits the edge into two edges. The edge closer to the (old) root of the tree gets a branch length of 0.0, and receives no placements. The edge further away from the root gets the branch length and all placements of the target_edge. See tree::make_rooted() for further details.
The function also recalculates the edge nums of all edges using reset_edge_nums(), and updates the placement data accordingly. This is because adding a node and an edge in the middle of the tree changes the traversal order, and hence the edge nums. It also takes care of changing the distal/proximal length values of the PqueryPlacements that sit in between the old (trifurcation) root and the new (proper) root.
Definition at line 51 of file placement/function/manipulation.cpp.
Returns a Sample where all Samples of a SampleSet have been merged into.
For this method to succeed, all Samples need to have the same topology, including identical edge_nums and node names. The Tree of the returned Sample has the average branch lenghts from the input trees, using TreeSet::average_branch_length_tree().
Definition at line 68 of file sample_set.cpp.
| void merge_duplicate_names | ( | Pquery & | pquery | ) |
Merge all PqueryNames that have the same name property into one, while adding up their multiplicity.
Definition at line 739 of file placement/function/functions.cpp.
| void merge_duplicate_names | ( | Sample & | smp | ) |
Call merge_duplicate_names() for each Pquery of the Sample.
Definition at line 760 of file placement/function/functions.cpp.
| void merge_duplicate_placements | ( | Pquery & | pquery | ) |
Merge all PqueryPlacements of a Pquery that are on the same TreeEdge into one averaged PqueryPlacement.
The merging is done via averaging all values of the PqueryPlacement: likelihood, like_weight_ratio, proximal_length, pendant_length and parsimony.
Definition at line 688 of file placement/function/functions.cpp.
| void merge_duplicate_placements | ( | Sample & | smp | ) |
Call merge_duplicate_placements( Pquery& ) for each Pquery of a Sample.
Definition at line 732 of file placement/function/functions.cpp.
| void merge_duplicates | ( | Sample & | smp | ) |
Look for Pqueries with the same name and merge them.
This function is a wrapper that simply calls three other functions on the provided Sample:
* collect_duplicate_pqueries() * merge_duplicate_names() * merge_duplicate_placements()
See there for more information on what they do.
Definition at line 569 of file placement/function/functions.cpp.
| NodeDistanceHistogramSet node_distance_histogram_set | ( | Sample const & | sample, |
| utils::Matrix< double > const & | node_distances, | ||
| utils::Matrix< signed char > const & | node_sides, | ||
| size_t const | histogram_bins | ||
| ) |
Calculate the NodeDistanceHistogramSet representing a single Sample, given the necessary matrices of this Sample.
This is a basic function that is mainly used for speedup in applications. The two matrices only depend on the tree, but not on the placement data, so for a set of Samples with the same tree, they only need to be calculated once.
|
static |
|
static |
|
static |
| double node_histogram_distance | ( | NodeDistanceHistogramSet const & | lhs, |
| NodeDistanceHistogramSet const & | rhs | ||
| ) |
| double node_histogram_distance | ( | Sample const & | sample_a, |
| Sample const & | sample_b, | ||
| size_t const | histogram_bins = 25 |
||
| ) |
Calculate the Node Histogram Distance of two Samples.
The necessary matrices of the Samples are calculated, then their NodeDistanceHistogramSet are build, and finally the distance is calculated. Basically, this is a high level function that simply chains node_distance_histogram_set() and node_histogram_distance() for convenience.
| utils::Matrix< double > node_histogram_distance | ( | SampleSet const & | sample_set, |
| size_t const | histogram_bins = 25 |
||
| ) |
| utils::Matrix< double > node_histogram_distance | ( | std::vector< NodeDistanceHistogramSet > const & | histogram_sets | ) |
| void normalize_weight_ratios | ( | Pquery & | pquery | ) |
Recalculate the like_weight_ratio of the PqueryPlacement&s of a Pquery, so that their sum is 1.0, while maintaining their ratio to each other.
Definition at line 123 of file placement/function/functions.cpp.
| void normalize_weight_ratios | ( | Sample & | smp | ) |
Recalculate the like_weight_ratio of the PqueryPlacement&s of each Pquery in the Sample, so that their sum is 1.0, while maintaining their ratio to each other.
This function simply calls normalize_weight_ratios( Pquery& pquery ) for all Pqueries of the Sample.
Definition at line 137 of file placement/function/functions.cpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| Sample const & | smp | ||
| ) |
Print a table of all Pqueries with their Placements and Names to the stream.
Definition at line 238 of file placement/function/operators.cpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| SampleSet const & | sample_set | ||
| ) |
Definition at line 181 of file sample_set.cpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| SimulatorEdgeDistribution const & | distrib | ||
| ) |
Definition at line 59 of file placement/simulator/functions.cpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| SimulatorExtraPlacementDistribution const & | distrib | ||
| ) |
Definition at line 66 of file placement/simulator/functions.cpp.
| std::ostream & operator<< | ( | std::ostream & | out, |
| SimulatorLikeWeightRatioDistribution const & | distrib | ||
| ) |
Definition at line 79 of file placement/simulator/functions.cpp.
| double pairwise_distance | ( | const Sample & | smp_a, |
| const Sample & | smp_b, | ||
| bool | with_pendant_length = false |
||
| ) |
Calculate the normalized pairwise distance between all placements of the two Samples.
This method calculates the distance between two Samples as the normalized sum of the distances between all pairs of Pqueries in the Sample. It is similar to the variance() calculation, which calculates this sum for the squared distances between all Pqueries of one Sample.
| smp_a | First Sample to which the distances shall be calculated to. |
| smp_b | Second Sample to which the distances shall be calculated to. |
| with_pendant_length | Whether or not to include all pendant lengths in the calculation. |
Definition at line 127 of file measures.cpp.
| std::vector< utils::Color > placement_color_count_gradient | ( | Sample const & | smp, |
| bool | linear | ||
| ) |
Returns a vector with a Color for each edge that visualizes the number of placements on that edge.
The vector is indexed using the edge.index(). Each edge gets assigned a Color value with these properties:
The gradient can be controlled via the linear parameter. If set to true, the scaling of the color gradient is linar in the number of placements. If set to false (default), it is logarithmic. This way, the color resolution is higher for low placement numbers, and compressed for higher numbers. A typical distribution of placements yields only some edges with a very high number of placements, while most of the other edges have little to no placements. Thus, it is reasonable to emphasize the differences between those edges with a lower placement count - which is what the default does.
See color heat_gradient() for more information.
Definition at line 71 of file placement/formats/edge_color.cpp.
| std::pair< PlacementTreeEdge const *, size_t > placement_count_max_edge | ( | Sample const & | smp | ) |
Get the number of placements on the edge with the most placements, and a pointer to this edge.
Definition at line 789 of file placement/function/functions.cpp.
| std::vector< size_t > placement_count_per_edge | ( | Sample const & | sample | ) |
Return a vector that contains the number of PqueryPlacements per edge of the tree of the Sample.
The vector is indexed using the index of the edges.
Definition at line 158 of file placement/function/helper.cpp.
| utils::Matrix< size_t > placement_count_per_edge | ( | SampleSet const & | sample_set | ) |
Definition at line 171 of file placement/function/helper.cpp.
| double placement_distance | ( | PqueryPlacement const & | place_a, |
| PqueryPlacement const & | place_b, | ||
| utils::Matrix< double > const & | node_distances | ||
| ) |
Calculate the distance between two PqueryPlacements, using their position on the tree::TreeEdges, measured in branch length units.
The Matrix node_distances has to come from tree::node_branch_length_distance_matrix().
Definition at line 157 of file placement/function/distances.cpp.
| double placement_distance | ( | PqueryPlacement const & | placement, |
| tree::TreeNode const & | node, | ||
| utils::Matrix< double > const & | node_distances | ||
| ) |
Calculate the distance in branch length units between a PqueryPlacement and a tree::TreeNode.
The Matrix node_distances has to come from tree::node_branch_length_distance_matrix().
Definition at line 287 of file placement/function/distances.cpp.
| std::pair< PlacementTreeEdge const *, double > placement_mass_max_edge | ( | Sample const & | smp | ) |
Get the summed mass of the placements on the heaviest edge, measured by their like_weight_ratio, and a pointer to this edge.
Definition at line 806 of file placement/function/functions.cpp.
| std::vector< double > placement_mass_per_edge_without_multiplicities | ( | Sample const & | sample | ) |
Return a vector that contains the sum of the masses of the PqueryPlacements per edge of the tree of the Sample.
The mass is measured in like_weight_ratio. The vector is indexed using the index of the edges.
Note that this does not take multiplicities into account; that is, it simply gives the sum of all like_weight_ratios. See placement_mass_per_edges_with_multiplicities() for a version of this function that also takes the multiplicities of the Pqueries into account.
Definition at line 164 of file masses.cpp.
| utils::Matrix< double > placement_mass_per_edge_without_multiplicities | ( | SampleSet const & | sample_set | ) |
Return a Matrix that contains the placement masses per edge.
See placement_mass_per_edge_without_multiplicities( Sample const& ) for details. The Matrix is simply a row-wise concatenation of this vector function.
Definition at line 177 of file masses.cpp.
| std::vector< double > placement_mass_per_edges_with_multiplicities | ( | Sample const & | sample | ) |
Return a vector that contains the sum of the masses of the PqueryPlacements per edge of the tree of the Sample, using the multiplicities as factors.
Definition at line 87 of file masses.cpp.
| utils::Matrix< double > placement_mass_per_edges_with_multiplicities | ( | SampleSet const & | sample_set | ) |
Return a Matrix that contains the placement masses per edge, using the multiplicities as factors.
See placement_mass_per_edges_with_multiplicities( Sample const& ) for details. The Matrix is simply a row-wise concatenation of this vector function.
Definition at line 101 of file masses.cpp.
| size_t placement_path_length_distance | ( | PqueryPlacement const & | placement, |
| tree::TreeEdge const & | edge, | ||
| utils::Matrix< size_t > const & | edge_path_lengths | ||
| ) |
Calculate the discrete distance from a PqueryPlacement to an edge, measured as the number of nodes between them.
The Matrix edge_path_lengths has to come from tree::edge_path_length_matrix().
Definition at line 342 of file placement/function/distances.cpp.
| size_t placement_path_length_distance | ( | PqueryPlacement const & | place_a, |
| PqueryPlacement const & | place_b, | ||
| utils::Matrix< size_t > const & | node_path_lengths | ||
| ) |
brief Calculate the discrete distance between two PqueryPlacements, using their position on the tree::TreeEdges, measured in number of nodes between the placement locations.
That is, two PqueryPlacements on the same edge have a distance of zero, on neighbouring edges a distance of 1 (as there is one node in between), and so on.
The Matrix node_path_lengths has to come from tree::node_path_length_matrix().
Definition at line 216 of file placement/function/distances.cpp.
| std::vector< std::vector< PqueryPlacement const * > > placements_per_edge | ( | Sample const & | smp, |
| bool | only_max_lwr_placements = false |
||
| ) |
Return a mapping from each PlacementTreeEdges to the PqueryPlacements that are placed on that edge.
The result vector is indexed using PlacementTreeEdge::index(). For each entry, it contains another vector that holds pointers to the PqueryPlacements of the Sample.
If the optional parameter only_max_lwr_placements is set to false (default), each placement in the Sample is added, not just the most likely ones. If set to true, only the PqueryPlacement with the highest like_weight_ratio is added.
The result is invalidated when calling Pquery::add_placement() or other functions that change the number of Pqueries or PqueryPlacements in the Sample.
Definition at line 106 of file placement/function/helper.cpp.
| std::vector< PqueryPlacement const * > placements_per_edge | ( | Sample const & | smp, |
| PlacementTreeEdge const & | edge | ||
| ) |
Return a vector of all PqueryPlacements that are placed on the given PlacementTreeEdge.
This functions iterates over all placements and collects those that are placed on the given edge. In case that this is needed for multiple edges, it is faster to use placements_per_edge( Sample ) instead.
The result is invalidated when calling Pquery::add_placement() or other functions that change the number of Pqueries or PqueryPlacements in the Sample.
Definition at line 141 of file placement/function/helper.cpp.
| std::vector< PqueryPlain > plain_queries | ( | Sample const & | smp | ) |
Return a plain representation of all pqueries of this map.
This method produces a whole copy of all pqueries and their placements (though, not their names) in a plain POD format. This format is meant for speeding up computations that need access to the data a lot - which would require several pointer indirections in the normal representation of the data.
This comes of course at the cost of reduced flexibility, as all indices are fixed in the plain data structre: changing a value here will not have any effect on the original data or even on the values of the pqueries. Thus, most probably this will lead to corruption. Therefore, this data structure is meant for reading only.
Definition at line 195 of file placement/function/helper.cpp.
| std::vector< std::vector< Pquery const * > > pqueries_per_edge | ( | Sample const & | sample, |
| bool | only_max_lwr_placements = false |
||
| ) |
Return a mapping from each edge to the Pqueries on that edge.
If only_max_lwr_placements is false (default), each PqueryPlacement of the Pqueries is counted. If true, only the most probable one is added to the map.
Definition at line 71 of file placement/function/helper.cpp.
| double pquery_distance | ( | Pquery const & | pquery, |
| tree::TreeNode const & | node, | ||
| utils::Matrix< double > const & | node_distances | ||
| ) |
Calculate the weighted distance between the PqueryPlacements of a Pquery and a tree::TreeNode, in branch length units, using the like_weight_ratio of the PqueryPlacements for weighing.
The Matrix node_distances has to come from tree::node_branch_length_distance_matrix().
Definition at line 274 of file placement/function/distances.cpp.
| double pquery_distance | ( | Pquery const & | pquery_a, |
| Pquery const & | pquery_b, | ||
| utils::Matrix< double > const & | node_distances, | ||
| bool | with_pendant_length = false |
||
| ) |
Calculate the weighted distance between two Pqueries, in branch length units, as the pairwise distance between their PqueryPlacements, and using the like_weight_ratio for weighing.
The Matrix node_distances has to come from tree::node_branch_length_distance_matrix().
Definition at line 138 of file placement/function/distances.cpp.
| double genesis::placement::pquery_distance | ( | Pquery const & | pquery, |
| DistanceFunction | distance_function | ||
| ) |
Local helper function to avoid code duplication.
Definition at line 255 of file placement/function/distances.cpp.
| double genesis::placement::pquery_distance | ( | Pquery const & | pquery_a, |
| Pquery const & | pquery_b, | ||
| DistanceFunction | distance_function | ||
| ) |
Local helper function to avoid code duplication.
Definition at line 112 of file placement/function/distances.cpp.
| double pquery_distance | ( | PqueryPlain const & | pquery_a, |
| PqueryPlain const & | pquery_b, | ||
| utils::Matrix< double > const & | node_distances, | ||
| bool | with_pendant_length = false |
||
| ) |
Calculate the weighted distance between two plain pqueries. It is mainly a helper method for distance calculations (e.g., pairwise distance, variance).
For each placement in the two pqueries, a distance is calculated, and their weighted sum is returned. Weighing is done using the mass of placements in both pqueries.
The distance between two placements is calculated as the shortest path between them. This includes the their position on the branches, and - if specified - the pendant_length of both. There are three cases that might occur:
The first case is easy to detect by comparing the edge nums. However, distinguishing between the latter two cases is expensive, as it involves finding the path to the root for both placements. To speed this up, we instead use a distance matrix that is calculated in the beginning of any algorithm using this method and contains the pairwise distances between all nodes of the tree. Using this, we do not need to find paths between placements, but simply go to the nodes at the end of the branches of the placements and do a lookup for those nodes.
With this technique, we can calculate the distances between the placements for all three cases (promixal-promixal, proximal-distal and distal-proximal) cheaply. The wanted distance is then simply the minimum of those three distances. This is correct, because the two wrong cases will always produce an overestimation of the distance.
This distance is normalized using the like_weight_ratio of both placements, before summing it up to calculate the total distance between the pqueries.
The Matrix node_distances has to come from tree::node_branch_length_distance_matrix().
Definition at line 51 of file placement/function/distances.cpp.
| double pquery_path_length_distance | ( | Pquery const & | pquery, |
| tree::TreeEdge const & | edge, | ||
| utils::Matrix< size_t > const & | edge_path_lengths | ||
| ) |
Calculate the weighted discrete distance between the PqueryPlacements of a Pquery and a tree::TreeEdge, in number of nodes between them, using the like_weight_ratio of the PqueryPlacements for weighing.
The Matrix edge_path_lengths has to come from tree::edge_path_length_matrix().
Definition at line 329 of file placement/function/distances.cpp.
| double pquery_path_length_distance | ( | Pquery const & | pquery_a, |
| Pquery const & | pquery_b, | ||
| utils::Matrix< size_t > const & | node_path_lengths | ||
| ) |
Calculate the weighted discrete distance between two Pqueries, measured as the pairwise distance in number of nodes between between their PqueryPlacements, and using the like_weight_ratio for weighing.
The Matrix node_path_lengths has to come from tree::node_path_length_matrix().
Definition at line 202 of file placement/function/distances.cpp.
| std::string print_tree | ( | Sample const & | smp | ) |
Return a simple view of the Tree of a Sample with information about the Pqueries on it.
Definition at line 275 of file placement/function/operators.cpp.
| void rectify_values | ( | Sample & | sample | ) |
Correct invalid values of the PqueryPlacements and PqueryNames as good as possible.
Some values can be slightly outside their valid boundaries, either for numerical reasons, or because something went wrong. Often, those can be rectified without too much loss of information.
This function
like_weight_ratio to 0.0like_weight_ratio > 1.0 to 1.0like_weight_ratio if their sum is > 1.0pendant_length to 0.0proximal_length to 0.0proximal_length > branch_length to branch_length for its edge.multiplicity to 0.0 for the nameSee rectify_values( SampleSet& ) for a version of this function that works on whole SampleSets.
Definition at line 229 of file placement/function/helper.cpp.
| void rectify_values | ( | SampleSet & | sset | ) |
Correct invalid values of the PqueryPlacements and PqueryNames as good as possible.
This function calls rectify_values( Sample& ) for all Samples in the SampleSet. See there for details.
Definition at line 277 of file placement/function/helper.cpp.
| size_t remove_empty_name_pqueries | ( | Sample & | sample | ) |
Remove all Pqueries from the Sample that have no PqueryNames.
This is useful for example after filtering, as this can result in removing all PqueryNames from a Pquery.
The function returns the number of removed Pqueries.
Definition at line 506 of file placement/function/functions.cpp.
| size_t remove_empty_placement_pqueries | ( | Sample & | sample | ) |
Remove all Pqueries from the Sample that have no PqueryPlacements.
This is useful for example after filtering, as this can result in removing all PqueryPlacements from a Pquery.
The function returns the number of removed Pqueries.
Definition at line 333 of file placement/function/functions.cpp.
| void reset_edge_nums | ( | PlacementTree & | tree | ) |
Reset all edge nums of a PlacementTree.
The edge_num property of the PlacementTreeEdges is defined by the jplace standard. The values have to be assigned increasingly with a postorder traversal of the tree. This function resets them so that this is established.
See has_correct_edge_nums() to check whether the edge nums are already correct. This should be the case for any valid jplace file.
Definition at line 285 of file placement/function/helper.cpp.
| void scale_all_branch_lengths | ( | Sample & | smp, |
| double | factor = 1.0 |
||
| ) |
Scale all branch lengths of the Tree and the position of the PqueryPlacements by a given factor.
This function calls tree::scale_all_branch_lengths() for scaling the tree, and also applies the same scaling to the PqueryPlacement::proximal_length.
Definition at line 168 of file placement/function/functions.cpp.
| void set_depths_distributed_weights | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of an SimulatorEdgeDistribution so that they follow the depth distribution of the edges in the provided Sample.
This function is similar to set_depths_distributed_weights( Sample const& sample, std::vector<int> const& depth_weights, SimulatorEdgeDistribution& edge_distrib ), but instead of using a given depth_weight vector, this vector is also estimated from the given Sample. This is done by using closest_leaf_weight_distribution(), which counts the number of placements at a given depth in the tree.
Definition at line 195 of file placement/simulator/functions.cpp.
| void set_depths_distributed_weights | ( | Sample const & | sample, |
| std::vector< double > const & | depth_weights, | ||
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights so that they follow a given depth distribution of the edges in the PlacementTree.
The depth_weights vector provides weights for each level of depth for an edge in the tree. This means, each edge which is adjacent to a leaf node (speak: it has depth 0) will use the weight at position 0; edges which are one level deeper in the tree will get the weight at position 1, and so on.
This method can conveniently be used with the output of closest_leaf_weight_distribution() called on some exemplary Sample. This way, it will mimic this sample in terms of the depths distribution of the placements: E.g., if the original sample (the one where the histrogram results were taken from and used as input for this method) has many placements near the leaves, so will the simulated one. See set_depths_distributed_weights( Sample const& sample, SimulatorEdgeDistribution& edge_distrib ) for a version of this function which does exaclty that.
Definition at line 218 of file placement/simulator/functions.cpp.
| void set_random_edges | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of a SimulatorEdgeDistribution randomly to either 0.0 or 1.0, so that a random subset of edges is selected (with the same probability for each selected edge).
The number of edges is taken from the provided Sample.
Definition at line 159 of file placement/simulator/functions.cpp.
| void set_random_edges | ( | size_t | edge_count, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of an SimulatorEdgeDistribution randomly to either 0.0 or 1.0, so that a random subset of edges is selected (with the same probability for each selected edge).
Definition at line 168 of file placement/simulator/functions.cpp.
| size_t set_random_subtree_weights | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Sets the weights of an SimulatorEdgeDistribution to 1.0 for a randomly chosen subtree, all others to 0.0.
Returns the index of the chosen edge.
Definition at line 265 of file placement/simulator/functions.cpp.
| void set_random_weights | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of an SimulatorEdgeDistribution for the edges randomly to a value between 0.0 and 1.0.
The number of edges is taken from the provided Sample.
Definition at line 130 of file placement/simulator/functions.cpp.
| void set_random_weights | ( | size_t | edge_count, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of an SimulatorEdgeDistribution for the edges randomly to a value between 0.0 and 1.0.
Definition at line 139 of file placement/simulator/functions.cpp.
| void set_subtree_weights | ( | Sample const & | sample, |
| size_t | link_index, | ||
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Set the weights of a subtree to 1.0 and all other weights to 0.0.
The subtree is selected via the index of the link that leads away from it. As leaf nodes do not count as subtrees, the link has to belong to an inner node.
Definition at line 312 of file placement/simulator/functions.cpp.
| void set_uniform_weights | ( | Sample const & | sample, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Sets the weights of an SimulatorEdgeDistribution to 1.0 for all edges, so that each edge has the same probability of being chosen.
The number of edges is taken from the provided Sample.
Definition at line 106 of file placement/simulator/functions.cpp.
| void set_uniform_weights | ( | size_t | edge_count, |
| SimulatorEdgeDistribution & | edge_distrib | ||
| ) |
Sets the weights of an SimulatorEdgeDistribution to 1.0 for all edges, so that each edge has the same probability of being chosen.
Definition at line 115 of file placement/simulator/functions.cpp.
| void sort_placements_by_weight | ( | Pquery & | pquery | ) |
Sort the PqueryPlacements of a Pquery by their like_weight_ratio, in descending order (most likely first).
Definition at line 147 of file placement/function/functions.cpp.
| void sort_placements_by_weight | ( | Sample & | smp | ) |
Sort the PqueryPlacements of all Pqueries by their like_weight_ratio, in descending order (most likely first).
Definition at line 161 of file placement/function/functions.cpp.
Definition at line 104 of file sample.cpp.
| double total_multiplicity | ( | Pquery const & | pqry | ) |
Return the sum of all multiplicities of the Pquery.
Definition at line 65 of file masses.cpp.
| double total_multiplicity | ( | Sample const & | sample | ) |
Return the sum of all multiplicities of all the Pqueries of the Sample.
Definition at line 74 of file masses.cpp.
| size_t total_name_count | ( | Sample const & | smp | ) |
Get the total number of PqueryNames in all Pqueries of the given Sample.
Definition at line 771 of file placement/function/functions.cpp.
| size_t total_placement_count | ( | Sample const & | smp | ) |
Get the total number of PqueryPlacements in all Pqueries of the given Sample.
Definition at line 780 of file placement/function/functions.cpp.
| double total_placement_mass_with_multiplicities | ( | Sample const & | smp | ) |
Get the mass of all PqueryPlacements of the Sample, using the multiplicities as factors.
This function returns the summed mass of all PqueryPlacements in all Pqueries of the given Sample, where mass is measured by like_weight_ratio, and the mass of each Pquery is multiplied by the sum of the multiplicities of this Pquery.
This method returns the same value as total_placement_mass_without_multiplicities() in case that the multiplicity is left at its default value of 1.0 for all Pqueries.
Definition at line 142 of file masses.cpp.
| double total_placement_mass_without_multiplicities | ( | Sample const & | smp | ) |
Get the summed mass of all PqueryPlacements in all Pqueries of the given Sample, where mass is measured by the like_weight_ratios of the PqueryPlacements.
Be aware that this function only gives the pure sum of the like_weight_ratios. See total_placement_mass_with_multiplicities() for a version of this function, which also takes the multiplicities of the Pqueries into account.
Definition at line 217 of file masses.cpp.
| size_t total_pquery_count | ( | SampleSet const & | sample_set | ) |
Return the total number of Pqueries in the Samples of the SampleSet.
Definition at line 107 of file sample_set.cpp.
| tree::TreeSet tree_set | ( | SampleSet const & | sample_set | ) |
Return a TreeSet containing all the trees of the SampleSet.
Definition at line 156 of file sample_set.cpp.
| bool validate | ( | Sample const & | smp, |
| bool | check_values = false, |
||
| bool | break_on_values = false |
||
| ) |
Validate the integrity of the pointers, references and data in a Sample object.
Returns true iff everything is set up correctly. In case of inconsistencies, the function stops and returns false on the first encountered error.
If check_values is set to true, also a check on the validity of numerical values is done, for example that the proximal_length is smaller than the corresponding branch_length. If additionally break_on_values is set, validate() will stop on the first encountered invalid value. Otherwise it will report all invalid values to the log stream.
Definition at line 343 of file placement/function/helper.cpp.
| double variance | ( | const Sample & | smp, |
| bool | with_pendant_length = false |
||
| ) |
Calculate the variance of the placements on a tree.
The variance is a measure of how far a set of items is spread out in its space (http://en.wikipedia.org/wiki/variance). In many cases, it can be measured using the mean of the items. However, when considering placements on a tree, this does not truly measure how far they are from each other. Thus, this algorithm applies a different method of calculating the variance in terms of squared deviations of all items from each other: \( Var(X) = \frac{1}{n^2} \sum_{i=1}^n \sum_{j=1}^n \frac{1}{2}(x_i - x_j)^2 \), where \( (x_i - x_j) \) denotes the distance between two placements.
According to the formula above, each pair of placements is evaluated twice, and subsequently their distance need to be halfed when being added to the sum of distanaces. Instead of that, we calculate the distance for each pair only once, thus are able skip half the calculations, and of course skip the division by two.
Furthermore, the normalizing factor \( \frac{1}{n^2} \) of the variance usually contains the number of elements being processed. However, as the placements are weighted by their like_weight_ratio, we instead calculate n as the sum of the like_weight_ratio of all placements. In case that for each pquery the ratios of all its placements sum up to 1.0, this number will be equal to the number of pqueries (and thus be equal to the usual case of using the number of elements). However, as this is not required (placements with small ratio can be dropped, so that their sum per pquery is less than 1.0), we cannout simply use the count.
Definition at line 195 of file measures.cpp.
|
static |
Internal function that calculates the sum of distances contributed by one pquery for the variance. See variance() for more information.
This function is intended to be called by variance() – it is not a stand-alone function.
Definition at line 173 of file measures.cpp.
| typedef tree::Tree PlacementTree |
Alias for a tree::Tree used for a tree with information needed for storing Pqueries. This kind of tree is used by Sample.
A PlacementTree inherits the data from tree::CommonTree, that is, it stores names for the nodes (usually those are taxa names) and branch lengths for the edges.
In addition to that, each edge of this tree has a value edge_num. This is not the same as the internally used index property of tree edges. Instead, it is a value defined by the jplace standard to identify edges. See Sample for more information.
Definition at line 54 of file placement/formats/edge_color.hpp.
| typedef tree::TreeEdge PlacementTreeEdge |
Alias for tree::TreeEdge used in a PlacementTree. See PlacementEdgeData for the data stored on the edges.
Definition at line 57 of file jplace_reader.hpp.
| using PlacementTreeLink = tree::TreeLink |
Alias for tree::TreeLink used in a PlacementTree.
Definition at line 74 of file placement_tree.hpp.
| using PlacementTreeNode = tree::TreeNode |
Alias for tree::TreeNode used in a PlacementTree. See PlacementNodeData for the data stored on the nodes.
Definition at line 63 of file placement_tree.hpp.